
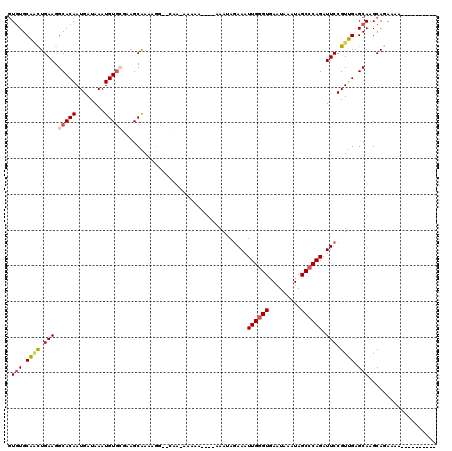
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,531,036 – 12,531,139 |
| Length | 103 |
| Max. P | 0.823839 |

| Location | 12,531,036 – 12,531,139 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.30 |
| Mean single sequence MFE | -21.97 |
| Consensus MFE | -16.17 |
| Energy contribution | -16.42 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823839 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
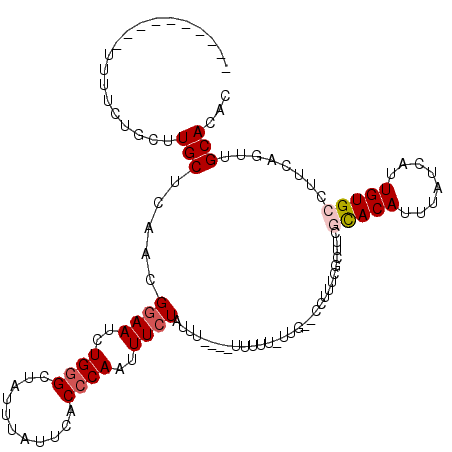
>3R_DroMel_CAF1 12531036 103 + 27905053 GUGUGCAUUUGAAGGCACAAUGAUAAAUGUACAAAGCAAAAGG--CAA-AGAAA----AAAUAGAAAUUGGGUGAAUAAAUAGCACAGAUUCCGUUGAGCAAGCAGAAAA---------- .......((((...((.(((((.............((.....)--)..-.....----.....(((.(((.((.........)).))).)))))))).))...))))...---------- ( -13.30) >DroVir_CAF1 13451 108 + 1 GUGUGCAGCUGAAGGCACAAUGAUAAAUGUGGUAAGCAAAAAC--GAAAAAAAAAAAGGAAAAGAAAUUGGGUGAAUAAAUAGCCCAGAUUCCGUUGAGCAAGCAGAAAA---------- ((.(((.(((...(.((((........)))).).)))......--..................(((.((((((.........)))))).)))......))).))......---------- ( -20.60) >DroPse_CAF1 19593 104 + 1 GUGUGCGACUGAAGGCACAAUGAUAAAUGUGCGGAGCGAAAGGAACAA-CAAAA----AAAUAGAAAUUGGGUGAAUAACUAGCCCAGAUUCCGUCGAGCAAGCAGAAA----------- .(((.((((.....(((((........)))))(((((....)......-.....----.........((((((.........)))))).)))))))).)))........----------- ( -26.00) >DroGri_CAF1 10964 104 + 1 GUGUGCAGCUGAAGGCACAAUGAUAAAUGUGGUAAGCAAAAAC--GAGCAGAAA----AAAAAUAAAUUGGGUGAAUAAAUAGCCCAGAUUCCGUUGAGCAAGCAGAAAA---------- ........(((...((.(((((.............((......--..)).....----.........((((((.........))))))....))))).))...)))....---------- ( -19.30) >DroAna_CAF1 19701 112 + 1 UUUUGCAACUGAAGGCACAAUGAUAAAUGUGCG-AGCGAAAGG--CAC-AAAAA----AAAUAGAAAUUGGGUGAAUAAAUAGCCCAGAUUCCGUUGAGCAAGCAAAAAAAAAAAAAGUA ((((((..(....)((.(((((.....(((((.-..(....))--)))-)....----.....(((.((((((.........)))))).)))))))).))..))))))............ ( -26.60) >DroPer_CAF1 23697 104 + 1 GUGUGCGACUGAAGGCACAAUGAUAAAUGUGCGGAGCGAAAGGAACAA-CAAAA----AAAUAGAAAUUGGGUGAAUAACUAGCCCAGAUUCCGUCGAGCAAGCAGAAA----------- .(((.((((.....(((((........)))))(((((....)......-.....----.........((((((.........)))))).)))))))).)))........----------- ( -26.00) >consensus GUGUGCAACUGAAGGCACAAUGAUAAAUGUGCGAAGCAAAAGG__CAA_AAAAA____AAAUAGAAAUUGGGUGAAUAAAUAGCCCAGAUUCCGUUGAGCAAGCAGAAAA__________ .(((.((((.(((.(((((........)))))...................................((((((.........)))))).))).)))).)))................... (-16.17 = -16.42 + 0.25)


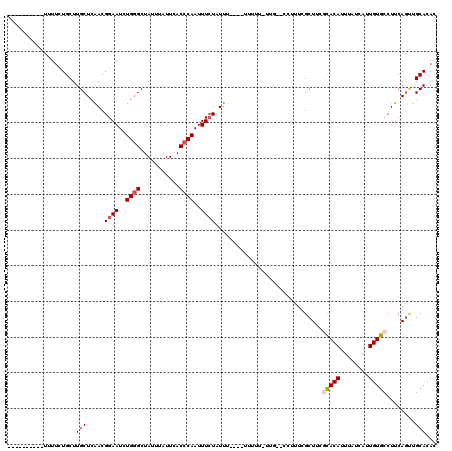
| Location | 12,531,036 – 12,531,139 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.30 |
| Mean single sequence MFE | -16.87 |
| Consensus MFE | -11.82 |
| Energy contribution | -12.35 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557531 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12531036 103 - 27905053 ----------UUUUCUGCUUGCUCAACGGAAUCUGUGCUAUUUAUUCACCCAAUUUCUAUUU----UUUCU-UUG--CCUUUUGCUUUGUACAUUUAUCAUUGUGCCUUCAAAUGCACAC ----------..(((((.((....)))))))..(((((.((((...................----.....-..(--(.....))...(((((........)))))....))))))))). ( -12.90) >DroVir_CAF1 13451 108 - 1 ----------UUUUCUGCUUGCUCAACGGAAUCUGGGCUAUUUAUUCACCCAAUUUCUUUUCCUUUUUUUUUUUC--GUUUUUGCUUACCACAUUUAUCAUUGUGCCUUCAGCUGCACAC ----------.....(((..(((....((((..((((...........))))..)))).................--............((((........)))).....))).)))... ( -14.80) >DroPse_CAF1 19593 104 - 1 -----------UUUCUGCUUGCUCGACGGAAUCUGGGCUAGUUAUUCACCCAAUUUCUAUUU----UUUUG-UUGUUCCUUUCGCUCCGCACAUUUAUCAUUGUGCCUUCAGUCGCACAC -----------........((..((((((((..((((...........))))..))))....----.....-................(((((........))))).....))))..)). ( -19.60) >DroGri_CAF1 10964 104 - 1 ----------UUUUCUGCUUGCUCAACGGAAUCUGGGCUAUUUAUUCACCCAAUUUAUUUUU----UUUCUGCUC--GUUUUUGCUUACCACAUUUAUCAUUGUGCCUUCAGCUGCACAC ----------.....(((..(((.((.((....((((...........))))..........----.....((..--......))....((((........)))))))).))).)))... ( -14.20) >DroAna_CAF1 19701 112 - 1 UACUUUUUUUUUUUUUGCUUGCUCAACGGAAUCUGGGCUAUUUAUUCACCCAAUUUCUAUUU----UUUUU-GUG--CCUUUCGCU-CGCACAUUUAUCAUUGUGCCUUCAGUUGCAAAA ............((((((..(((....((((..((((...........))))..))))....----.....-(.(--(.....)).-)(((((........)))))....))).)))))) ( -20.10) >DroPer_CAF1 23697 104 - 1 -----------UUUCUGCUUGCUCGACGGAAUCUGGGCUAGUUAUUCACCCAAUUUCUAUUU----UUUUG-UUGUUCCUUUCGCUCCGCACAUUUAUCAUUGUGCCUUCAGUCGCACAC -----------........((..((((((((..((((...........))))..))))....----.....-................(((((........))))).....))))..)). ( -19.60) >consensus __________UUUUCUGCUUGCUCAACGGAAUCUGGGCUAUUUAUUCACCCAAUUUCUAUUU____UUUUU_UUG__CCUUUCGCUUCGCACAUUUAUCAUUGUGCCUUCAGUUGCACAC ...................(((.....((((..((((...........))))..))))..............................(((((........)))))........)))... (-11.82 = -12.35 + 0.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:09 2006