| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,525,341 – 12,525,450 |
| Length | 109 |
| Max. P | 0.561587 |

| Location | 12,525,341 – 12,525,450 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.07 |
| Mean single sequence MFE | -22.94 |
| Consensus MFE | -18.38 |
| Energy contribution | -18.72 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.561587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
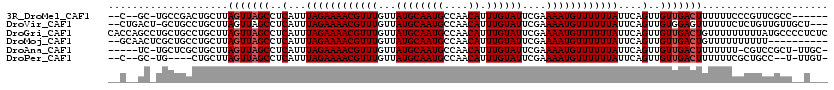
>3R_DroMel_CAF1 12525341 109 - 27905053 --C--GC-UGCCGACUGCUUAGUUAGCCUCAUUUAGAAAACGUUUGUUAUGCAAUGCCAACAUUUGUAUUCGAAAAUGUUUUUUAUUCAGUUGUUGACUUUUUUCCCGUUCGCC------ --.--..-.((.(((.....(((((((..(...((((((((((((...((((((((....)).))))))....))))))))))))....)..)))))))........))).)).------ ( -22.02) >DroVir_CAF1 6790 114 - 1 --CUGACU-GCUGCCUGCUUAGUUAGCCUCAUUUAGAAAACGUUUGUUAUGCAAUGCCAACAUUUGUAUUCGAAAAUGUUUUUUAUUCAGUUGUGGAGUUUUUUCUCUGUUGUUGCU--- --((((((-((.....))..)))))).......((((((((((((...((((((((....)).))))))....))))))))))))..(((..(..(((......)))..)..)))..--- ( -24.40) >DroGri_CAF1 3887 120 - 1 CACCAGCCUGCUGCCUGCUUAGUUAGCCUCAUUUAGAAAACGUUUGUUAUGCAAUGCCAACAUUUGUAUUCGAAAAUGUUUUUUAUUCAGUUGUUGACUGUUUUUUUUUAUGCCCCUCUC ...(((....)))......((((((((..(...((((((((((((...((((((((....)).))))))....))))))))))))....)..)))))))).................... ( -22.70) >DroMoj_CAF1 7261 108 - 1 --GCAACUCGCUGCCUGCUUAGUUAGCCUCAUUUAGAAAACGUUUGUUAUGCAAUGCCAACAUUUGUAUUCGAAAAUGUUUUUUAUUCAGUUGUUGACUGUUUUUUUUUU---------- --(((......))).....((((((((..(...((((((((((((...((((((((....)).))))))....))))))))))))....)..))))))))..........---------- ( -22.40) >DroAna_CAF1 14579 111 - 1 -----UC-UGCUCGCUGCUUAGUUAGCCUCAUUUAGAAAACGUUUGUUAUGCAAUGCCAACAUUUGUAUUCGAAAAUGUUUUUUAUUCAGUUGUUGACUUUUUUU-CGUCCGCU-UUGC- -----..-.((.((......(((((((..(...((((((((((((...((((((((....)).))))))....))))))))))))....)..)))))))......-))...)).-....- ( -22.00) >DroPer_CAF1 17481 107 - 1 --C--GC-UG----CUGCUUAGUUAGCCUCAUUUAGAAAACGUUUGUUAUGCAAUGCCAACAUUUGUAUUCGAAAAUGUUUUUUAUUCAGUUGUUGACUUUUUUCGCUGCC--U-UUGU- --.--((-.(----(.....(((((((..(...((((((((((((...((((((((....)).))))))....))))))))))))....)..)))))))......)).)).--.-....- ( -24.10) >consensus __C__GC_UGCUGCCUGCUUAGUUAGCCUCAUUUAGAAAACGUUUGUUAUGCAAUGCCAACAUUUGUAUUCGAAAAUGUUUUUUAUUCAGUUGUUGACUUUUUUCUCUUUCGCU__U___ ....................(((((((..(...((((((((((((...((((((((....)).))))))....))))))))))))....)..)))))))..................... (-18.38 = -18.72 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:03 2006