| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
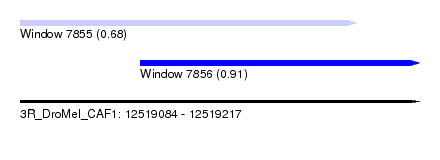
| Location | 12,519,084 – 12,519,217 |
| Length | 133 |
| Max. P | 0.913652 |

| Location | 12,519,084 – 12,519,196 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.89 |
| Mean single sequence MFE | -20.79 |
| Consensus MFE | -16.67 |
| Energy contribution | -16.20 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.679900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12519084 112 + 27905053 UCGAAAUAUUAUUAAGUCCCAAAGCUGCACUUCAAUUAAAAUGCAUUUUCGCAGUUUAUGGGCUUAAUCUCAUAAACAAUUUCUGCAUUGUUGCAAAUUCGCUCUCGUUCCU-------- .((((.....((((((.(((((((((((......................))))))).))))))))))...............((((....))))..))))...........-------- ( -22.95) >DroPse_CAF1 6860 117 + 1 UCGAAAUAUUAUAAAGUCCCAAAGCUGCACUUCAAUUAAAAUGCAUUUUCGCAGUUUAUGAGCUUAAUCUCAUAAACAAUUUCUGUAUUGUUGCGUC---UCCCUCAACCCUCACCCCUC .......................((.(((...((.......(((......)))(((((((((......)))))))))......))...))).))...---.................... ( -16.90) >DroEre_CAF1 5716 112 + 1 UCGAAAUAUUAUUAAGUCCCAAAGCUGCACUUCAAUUAAAAUGCAUUUUCGCAGUUUAUGGGCUUAAUCUCAUAAACAAUUUCUGCAUUGUUGCAAAUUCGCUCUCCUUCCU-------- .((((.....((((((.(((((((((((......................))))))).))))))))))...............((((....))))..))))...........-------- ( -22.95) >DroYak_CAF1 4957 112 + 1 UCGAAAUAUUAUUAAGUCCCAAAGCUGCACUUCAAUUAAAAUGCAUUUUCGCAGUUUAUGGGCUUAAUCUCAUAAACAAUUUCAGCAUUGUUGCAAAUUUGCUCUCCGCCCU-------- ..........((((((.(((((((((((......................))))))).))))))))))......((((((......))))))(((....)))..........-------- ( -21.05) >DroAna_CAF1 8877 110 + 1 UCGAAAUAUUAUUAAGUCCCAAAGCUGCACUUCAAUUAAAAUGCAUUUUCACAGUUUAUGAGCUUAAUCUCAUAAACAAUUUCUGCAUUGUGGCCGAGUGUUUCUUUUU--G-------- ..((((((((.....(((.(((.((((((.((......)).))))........(((((((((......))))))))).......)).))).)))..)))))))).....--.-------- ( -24.00) >DroPer_CAF1 10820 107 + 1 UCGAAAUAUUAUAAAGUCCCAAAGCUGCACUUCAAUUAAAAUGCAUUUUCGCAGUUUAUGAGCUUAAUCUCAUAAACAAUUUCUGUAUUGUUGCGUC---UCCCUCACCC---------- .......................((.(((...((.......(((......)))(((((((((......)))))))))......))...))).))...---..........---------- ( -16.90) >consensus UCGAAAUAUUAUUAAGUCCCAAAGCUGCACUUCAAUUAAAAUGCAUUUUCGCAGUUUAUGAGCUUAAUCUCAUAAACAAUUUCUGCAUUGUUGCAAAUU_GCUCUCAUCCCU________ ...............((..(((.((((((.((......)).))))........(((((((((......))))))))).......)).)))..)).......................... (-16.67 = -16.20 + -0.47)



| Location | 12,519,124 – 12,519,217 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 77.96 |
| Mean single sequence MFE | -19.85 |
| Consensus MFE | -12.53 |
| Energy contribution | -12.50 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12519124 93 + 27905053 AUGCAUUUUCGCAGUUUAUGGGCUUAAUCUCAUAAACAAUUUCUGCAUUGUUGCAAAUUCGCUCUCGUUCCUCGA-CGUUUUGGC--------AUU---------GUUGAU .(((......)))(((((((((......)))))))))......((((....)))).....(((..((((....))-))....)))--------...---------...... ( -19.70) >DroSec_CAF1 8155 93 + 1 AUGCAUUUUCGCAGUUUAUGGGCUUAAUCUCAUAAACAAUUUCUGCAUUGUUGCAAAUUCGCUCUCCUUCCUAGA-CUUUUUGGC--------AUU---------GUUGAU .(((......)))(((((((((......)))))))))....((.(((.(((((.(((.((.............))-.))).))))--------).)---------)).)). ( -17.32) >DroSim_CAF1 7927 93 + 1 AUGCAUUUUCGCAGUUUAUGGGCUUAAUCUCAUAAACAAUUUCUGCAUUGUUGCAAAUUCGCUCUCCUUCCUCGA-CUUUUUGGC--------AUU---------GUUGAU .(((......)))(((((((((......)))))))))....((.(((.(((((.(((.(((...........)))-.))).))))--------).)---------)).)). ( -18.20) >DroYak_CAF1 4997 94 + 1 AUGCAUUUUCGCAGUUUAUGGGCUUAAUCUCAUAAACAAUUUCAGCAUUGUUGCAAAUUUGCUCUCCGCCCUCGGAUUUUUUGCC--------AUU---------GUUGAU .(((......)))(((((((((......)))))))))....((((((.((..(((((.......((((....))))...))))))--------).)---------))))). ( -24.50) >DroAna_CAF1 8917 88 + 1 AUGCAUUUUCACAGUUUAUGAGCUUAAUCUCAUAAACAAUUUCUGCAUUGUGGCCGAGUGUUUCUUUUU--GAAG-CAUAUUUGC--------------------GUUGAU (((((........(((((((((......)))))))))....((.((......)).))(((((((.....--))))-)))...)))--------------------)).... ( -21.90) >DroPer_CAF1 10860 105 + 1 AUGCAUUUUCGCAGUUUAUGAGCUUAAUCUCAUAAACAAUUUCUGUAUUGUUGCGUC---UCCCUCACCC--CCA-CUUAUUGGCUUUUUGCUACUUGAAUUCGCUCUGAU ..((.....(((((((((((((......)))))))))(((......)))..))))..---..........--...-.....((((.....)))).........))...... ( -17.50) >consensus AUGCAUUUUCGCAGUUUAUGGGCUUAAUCUCAUAAACAAUUUCUGCAUUGUUGCAAAUUCGCUCUCCUUCCUCGA_CUUUUUGGC________AUU_________GUUGAU .(((......)))(((((((((......))))))))).......(((....)))......................................................... (-12.53 = -12.50 + -0.03)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:56 2006