| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,516,307 – 12,516,428 |
| Length | 121 |
| Max. P | 0.829825 |

| Location | 12,516,307 – 12,516,403 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.48 |
| Mean single sequence MFE | -31.63 |
| Consensus MFE | -19.58 |
| Energy contribution | -19.83 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.758389 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12516307 96 + 27905053 UAAAGCAACA--CGCGGCGCAACAUGCA---------------------ACGUGCAACAUGCAACAUGCGAGUUGGCAUUGCACGUUCCGGGAAUAGGAAUGGAAAUGGCUGCGGA-CUG ..........--((((((.((((.((((---------------------...(((.....)))...)))).)))).((((...((((((.......))))))..))))))))))..-... ( -34.00) >DroPse_CAF1 3601 112 + 1 UAAAGCAACA--CGCGGUGCAACAUGCAGCGUGCAGCAUGCAGCAGGCAGCAUGCAGCAUGCACCAUGCGAGCUGCCAUUGCACGUUCCGGGAAUAGGAAUGGAAAUGG-----GA-CUG ...(((....--((((((((.....)).((((((.((((((........)))))).)))))))))..))).))).(((((...((((((.......))))))..)))))-----..-... ( -42.70) >DroSec_CAF1 5338 96 + 1 UAAAGCAACA--CGCGGCGCAACAUGCA---------------------AAGUGCAACAUGCAACAUGCGAGUUGCCAUUGCACGUUCCGGGAAUAGGAAUGGAAAUGGCUUUGGA-CUG ((((((....--.((((.(((((.((((---------------------...(((.....)))...)))).)))))..)))).((((((.......))))))......))))))..-... ( -31.30) >DroYak_CAF1 2180 96 + 1 UAAAGCAACA--CGCGGCGCAACAUGCA---------------------ACGUGCAACAUGCAACAUGCGAGCUGCCAUUGCACGUUCCGGGAAUAGGAAUGGAAAUGGCUAUGCA-AUG ....(((...--.((..((((...((((---------------------..........))))...)))).)).((((((...((((((.......))))))..))))))..))).-... ( -30.70) >DroAna_CAF1 6275 92 + 1 UAAAGCAACACACGCGGUGCAACAUG----------------------------CAACAUGCAACAUGCGAGCUGCCAUUGCACGUUCCGGGAAUAGGAAUGGAAAUGGAAAUGGGAAAG .............((..((((...((----------------------------(.....)))...)))).))..(((((...((((((.......))))))..)))))........... ( -22.50) >DroPer_CAF1 7587 91 + 1 UAAAGCAACA--CGCGGUGCAACAUGCA---------------------GCAUGCAGCAUGCAACAUGCGAGCUGCCAUUGCACGUUCCGGGAAUAGGAAUGGAAAUGG-----AA-AUG ...(((....--((((.((((...(((.---------------------....)))...))))...)))).))).(((((...((((((.......))))))..)))))-----..-... ( -28.60) >consensus UAAAGCAACA__CGCGGCGCAACAUGCA_____________________ACGUGCAACAUGCAACAUGCGAGCUGCCAUUGCACGUUCCGGGAAUAGGAAUGGAAAUGGCU_UGGA_AUG ............((((..(((...((((........................))))...)))....)))).....(((((...((((((.......))))))..)))))........... (-19.58 = -19.83 + 0.25)



| Location | 12,516,307 – 12,516,403 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.48 |
| Mean single sequence MFE | -30.39 |
| Consensus MFE | -21.19 |
| Energy contribution | -21.05 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829825 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12516307 96 - 27905053 CAG-UCCGCAGCCAUUUCCAUUCCUAUUCCCGGAACGUGCAAUGCCAACUCGCAUGUUGCAUGUUGCACGU---------------------UGCAUGUUGCGCCGCG--UGUUGCUUUA (((-..(((.((((.......(((.......)))((((((((((.((((..((.....))..))))..)))---------------------))))))))).)).)))--..)))..... ( -29.40) >DroPse_CAF1 3601 112 - 1 CAG-UC-----CCAUUUCCAUUCCUAUUCCCGGAACGUGCAAUGGCAGCUCGCAUGGUGCAUGCUGCAUGCUGCCUGCUGCAUGCUGCACGCUGCAUGUUGCACCGCG--UGUUGCUUUA ...-..-----.((((.((.((((.......)))).).).))))(((((.(((..((((((....((((((.((.(((........))).)).)))))))))))))))--.))))).... ( -41.70) >DroSec_CAF1 5338 96 - 1 CAG-UCCAAAGCCAUUUCCAUUCCUAUUCCCGGAACGUGCAAUGGCAACUCGCAUGUUGCAUGUUGCACUU---------------------UGCAUGUUGCGCCGCG--UGUUGCUUUA ...-...(((((........((((.......)))).(((((((((((((......))))).))))))))..---------------------.((((((......)))--))).))))). ( -27.20) >DroYak_CAF1 2180 96 - 1 CAU-UGCAUAGCCAUUUCCAUUCCUAUUCCCGGAACGUGCAAUGGCAGCUCGCAUGUUGCAUGUUGCACGU---------------------UGCAUGUUGCGCCGCG--UGUUGCUUUA (((-(((((..((..................))...))))))))(((((.(((.(((.((((((.......---------------------.)))))).)))..)))--.))))).... ( -31.27) >DroAna_CAF1 6275 92 - 1 CUUUCCCAUUUCCAUUUCCAUUCCUAUUCCCGGAACGUGCAAUGGCAGCUCGCAUGUUGCAUGUUG----------------------------CAUGUUGCACCGCGUGUGUUGCUUUA ...............................(.((((((((((((((((......))))).)))))----------------------------)))))).)...(((.....))).... ( -23.80) >DroPer_CAF1 7587 91 - 1 CAU-UU-----CCAUUUCCAUUCCUAUUCCCGGAACGUGCAAUGGCAGCUCGCAUGUUGCAUGCUGCAUGC---------------------UGCAUGUUGCACCGCG--UGUUGCUUUA ...-..-----.((((.((.((((.......)))).).).))))(((((.(((.(((.((((((.......---------------------.)))))).)))..)))--.))))).... ( -29.00) >consensus CAG_UCCA_AGCCAUUUCCAUUCCUAUUCCCGGAACGUGCAAUGGCAGCUCGCAUGUUGCAUGUUGCACGU_____________________UGCAUGUUGCACCGCG__UGUUGCUUUA ............((((.((.((((.......)))).).).))))(((((.(((.(((.((((((.............................)))))).)))..)))...))))).... (-21.19 = -21.05 + -0.14)


| Location | 12,516,333 – 12,516,428 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 84.28 |
| Mean single sequence MFE | -22.40 |
| Consensus MFE | -17.97 |
| Energy contribution | -18.38 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.525503 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12516333 95 - 27905053 CCAUUCCAGUU-----CCCAAUCCCAGCCCCAGUCCGCAGCCAUUUCCAUUCCUAUUCCCGGAACGUGCAAUGCCAACUCGCAUGUUGCAUGUUGCACGU--------- ...........-----..........((........))............(((.......)))(((((((((..((((......))))...)))))))))--------- ( -17.60) >DroPse_CAF1 3639 104 - 1 GGGUUCAAGUUGCAGUCCCAGUGCCAGUCCCAGUC-----CCAUUUCCAUUCCUAUUCCCGGAACGUGCAAUGGCAGCUCGCAUGGUGCAUGCUGCAUGCUGCCUGCUG ((((...(((((((((....(..(((...(.(((.-----(((((.((.((((.......)))).).).)))))..))).)..)))..)..)))))).))))))).... ( -29.50) >DroSec_CAF1 5364 95 - 1 CCAUUCCAGUU-----CCCAAUCCCAGCCGCAGUCCAAAGCCAUUUCCAUUCCUAUUCCCGGAACGUGCAAUGGCAACUCGCAUGUUGCAUGUUGCACUU--------- ........(((-----(((.......)..((........))...................)))))(((((((((((((......))))).))))))))..--------- ( -20.70) >DroSim_CAF1 5461 95 - 1 CCAUUCCAGUU-----CCCAAUCCCAGCCCCAGUCCACAGCCAUUUCCAUUCCUAUUCCCGGAACGUGCAAUGGCAACUCGCAUGUUGCAUGUUGCACGU--------- ...........-----..................................(((.......)))(((((((((((((((......))))).))))))))))--------- ( -21.40) >DroEre_CAF1 2960 90 - 1 -----CCAGUU-----CCCAAUUCCAGCUCCAUUCCACAGCCAUUUCCAUUCCUAUUCCCGGAACGUGCAAUGGCAACUCGCAUGUUGCAUGUUGCACGU--------- -----......-----..........(((.........))).........(((.......)))(((((((((((((((......))))).))))))))))--------- ( -21.60) >DroYak_CAF1 2206 95 - 1 CCAUUCCAGUU-----CCCAACUCCAGCCCCAUUGCAUAGCCAUUUCCAUUCCUAUUCCCGGAACGUGCAAUGGCAGCUCGCAUGUUGCAUGUUGCACGU--------- ........(((-----((........((......))((((............))))....)))))(((((((((((((......))))).))))))))..--------- ( -23.60) >consensus CCAUUCCAGUU_____CCCAAUCCCAGCCCCAGUCCACAGCCAUUUCCAUUCCUAUUCCCGGAACGUGCAAUGGCAACUCGCAUGUUGCAUGUUGCACGU_________ .................................................((((.......))))((((((((((((((......))))).))))))))).......... (-17.97 = -18.38 + 0.42)

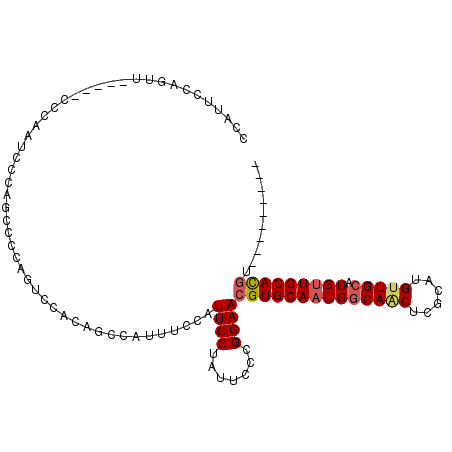

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:54 2006