
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,515,396 – 12,515,504 |
| Length | 108 |
| Max. P | 0.662651 |

| Location | 12,515,396 – 12,515,504 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.14 |
| Mean single sequence MFE | -23.65 |
| Consensus MFE | -20.00 |
| Energy contribution | -20.28 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.662651 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
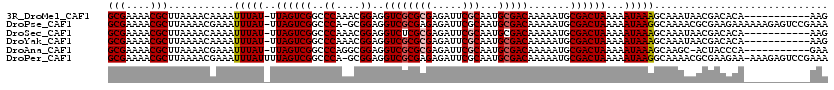
>3R_DroMel_CAF1 12515396 108 - 27905053 GCGAAAACGCUUAAAACAAAAUUUAU-UUAGUCGGCCCAAACGGAGGUCGCGCGAGAUUCGCAAUGCGACAAAAAUGCGACUAAAAAUAAAGCAAAUAACGACACA-----------AAG (((....)))...............(-(((((((..((....))..((((((((.....)))...))))).......)))))))).....................-----------... ( -24.00) >DroPse_CAF1 2558 118 - 1 GCGAAAACGCUUAAAACGAAAUUUAU-UUAGUCGGCCCA-GCGGAGGUCGCGAGAGAUUCGCAAUGCGACAAAAAUGCGACUAAAAAUAAGGCAAAACGCGAAGAAAAAAGAGUCCGAAA (((....)))......((.......(-(((((((((...-(((((..((....))..)))))...))..((....))))))))))......((.....))...............))... ( -24.10) >DroSec_CAF1 4414 108 - 1 GCGAAAACGCUUAAAACAAAAUUUAU-UUAGUCGGCCCAAACGGAGGUCUCGCGAGAUUCGCAAUGCGACAAAAAUGCGACUAAAAAUAAAGCAAAUAACGACACA-----------AAG (((....)))...............(-((((((((((........))))..(((....((((...))))......)))))))))).....................-----------... ( -18.90) >DroYak_CAF1 1316 108 - 1 GCGAAAACGCUUAAAACAAAAUUUAU-UUAGUCGGCCCAAACGGAGGUCGCGCGAGAUUCGCAAUGCGACAAAAAUGCGACUAAAAAUAAAGCAAAUAACGACACA-----------AAG (((....)))...............(-(((((((..((....))..((((((((.....)))...))))).......)))))))).....................-----------... ( -24.00) >DroAna_CAF1 5478 107 - 1 GCGAAAACGCUUAAAACGAAAUUUAU-UUAGUCGGCCCAGGCGGAGGUCGCGCGAGAUUCGCAAUGCGACAAAAAUGCGACUAAAAAUAAAGCAAGC-ACUACCCA-----------GAA (((....)))...............(-(((((((((....))....((((((((.....)))...))))).......))))))))............-........-----------... ( -25.80) >DroPer_CAF1 6477 118 - 1 GCGAAAACGCUUAAAACGAAAUUUAUUUUAGUCGGCCCA-GCGGAGGUCGCGAGAGAUUCGCAAUGCGACAAAAAUGCGACUAAAAAUAAGGCAAAACGCGAAGAA-AAAGAGUCCGAAA (((....)))......((...(((((((((((((((...-(((((..((....))..)))))...))..((....))))))).))))))))((.....))......-........))... ( -25.10) >consensus GCGAAAACGCUUAAAACAAAAUUUAU_UUAGUCGGCCCAAACGGAGGUCGCGCGAGAUUCGCAAUGCGACAAAAAUGCGACUAAAAAUAAAGCAAAUAACGACACA___________AAA (((....)))...........(((((..((((((..((....))..((((((((.....)))...))))).......))))))...)))))............................. (-20.00 = -20.28 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:52 2006