
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,512,973 – 12,513,102 |
| Length | 129 |
| Max. P | 0.984072 |

| Location | 12,512,973 – 12,513,066 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.47 |
| Mean single sequence MFE | -30.48 |
| Consensus MFE | -22.89 |
| Energy contribution | -21.83 |
| Covariance contribution | -1.05 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.984072 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
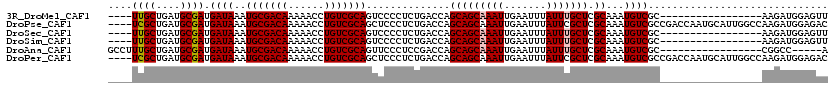
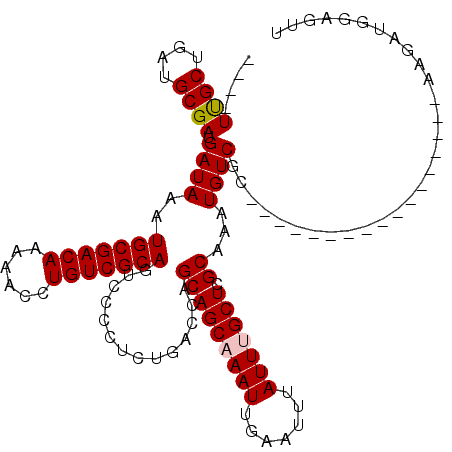
>3R_DroMel_CAF1 12512973 93 + 27905053 UACCUUUUCUGUUGUUCCCGU----------UUUGGCCUUAACUCCAUCUU-----------------GCGACAUUUGCGAGCAAAUAAAUUCAAUUUGCUGCUGGUCAGAGGGGACUGC ..........((.((((((..----------(((((((............(-----------------((((...)))))(((((((.......)))))))...))))))))))))).)) ( -25.10) >DroPse_CAF1 97 104 + 1 ---GUUUUC---UGUUCCCAU----------UUUGGCUGUGUCUCCAUCUUGGCCAAUGCAUUGGUCGGCGACAUUUGCGAGCGAAUAAAUUCAAUUUGCUGCUGGUCAGAGGGAGCUGC ---......---.((((((..----------((((((((((((.((.....((((((....)))))))).)))))..((.(((((((.......))))))))).)))))))))))))... ( -35.80) >DroSec_CAF1 2069 93 + 1 UACCUUUUCUGUUGUUCCCGU----------UUUGGCCUUAACUCCAUCUU-----------------GCGACAUUUGCGAGCAAAUAAAUUCAAUUUGCUGCUGGUCAGAGGGGACUGC ..........((.((((((..----------(((((((............(-----------------((((...)))))(((((((.......)))))))...))))))))))))).)) ( -25.10) >DroSim_CAF1 2134 93 + 1 UACCUUUUCUGUUGUUCCCGU----------UUUGGCCUUAACUCCAUCUU-----------------GCGACAUUUGCGAGCAAAUAAAUUCAAUUUGCUGCUGGUCAGAGGGGACUGC ..........((.((((((..----------(((((((............(-----------------((((...)))))(((((((.......)))))))...))))))))))))).)) ( -25.10) >DroAna_CAF1 3625 93 + 1 ---C--CGAUGUUGUUCCCGUUUUUGGCCGUUUUGGCCAUU-----GGCCG-----------------GCGACAUUUGCGAGCAAAUAAAUUCAAUUUGCUGCUGGUCGGAGGGAACUGC ---.--....((.((((((.....(((((.....)))))((-----(((((-----------------(((.(....)).(((((((.......)))))))))))))))).)))))).)) ( -36.00) >DroPer_CAF1 3909 104 + 1 ---GUUUUC---UGUUCCCAU----------UUUGGCUGUGUCUCCAUCUUGGCCAAUGCAUUGGUCGGCGACAUUUGCGAGCGAAUAAAUUCAAUUUGCUGCUGGUCAGAGGGAGCUGC ---......---.((((((..----------((((((((((((.((.....((((((....)))))))).)))))..((.(((((((.......))))))))).)))))))))))))... ( -35.80) >consensus ___CUUUUCUGUUGUUCCCGU__________UUUGGCCUUAACUCCAUCUU_________________GCGACAUUUGCGAGCAAAUAAAUUCAAUUUGCUGCUGGUCAGAGGGAACUGC .............((((((............(((((((..............................((((...)))).(((((((.......)))))))...)))))))))))))... (-22.89 = -21.83 + -1.05)



| Location | 12,513,003 – 12,513,102 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.56 |
| Mean single sequence MFE | -33.37 |
| Consensus MFE | -27.53 |
| Energy contribution | -26.83 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795738 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12513003 99 + 27905053 AACUCCAUCUU-----------------GCGACAUUUGCGAGCAAAUAAAUUCAAUUUGCUGCUGGUCAGAGGGGACUGCGACAGGUUUUUGUCGCAUUUAUCAUCGCAUCAGCAA---- ..((((.(((.-----------------..(((....((.(((((((.......)))))))))..))))))))))..((((((((....)))))))).........((....))..---- ( -28.90) >DroPse_CAF1 121 116 + 1 GUCUCCAUCUUGGCCAAUGCAUUGGUCGGCGACAUUUGCGAGCGAAUAAAUUCAAUUUGCUGCUGGUCAGAGGGAGCUGCGACAGGUUUUUGUCGCAUUUAUCAUCGCAUCAGCGA---- (.((((.(((.(((((.((((...(((...)))...))))(((((((.......)))))))..)))))))).)))))((((((((....)))))))).......((((....))))---- ( -40.10) >DroSec_CAF1 2099 99 + 1 AACUCCAUCUU-----------------GCGACAUUUGCGAGCAAAUAAAUUCAAUUUGCUGCUGGUCAGAGGGGACUGCGACAGGUUUUUGUCGCAUUUAUCAUCGCAUCAGCAA---- ..((((.(((.-----------------..(((....((.(((((((.......)))))))))..))))))))))..((((((((....)))))))).........((....))..---- ( -28.90) >DroSim_CAF1 2164 99 + 1 AACUCCAUCUU-----------------GCGACAUUUGCGAGCAAAUAAAUUCAAUUUGCUGCUGGUCAGAGGGGACUGCGACAGGUUUUUGUCGCAUUUAUCAUCGCAUCAGCAA---- ..((((.(((.-----------------..(((....((.(((((((.......)))))))))..))))))))))..((((((((....)))))))).........((....))..---- ( -28.90) >DroAna_CAF1 3660 98 + 1 U-----GGCCG-----------------GCGACAUUUGCGAGCAAAUAAAUUCAAUUUGCUGCUGGUCGGAGGGAACUGCGACAGGUUUUUGUCGCAUUUAUCAUCGCAUCAGCAAAGGC .-----.(((.-----------------((((...)))).(((((((.......)))))))((((((((..(..((.((((((((....)))))))).))..)..)).))))))...))) ( -33.30) >DroPer_CAF1 3933 116 + 1 GUCUCCAUCUUGGCCAAUGCAUUGGUCGGCGACAUUUGCGAGCGAAUAAAUUCAAUUUGCUGCUGGUCAGAGGGAGCUGCGACAGGUUUUUGUCGCAUUUAUCAUCGCAUCAGCGA---- (.((((.(((.(((((.((((...(((...)))...))))(((((((.......)))))))..)))))))).)))))((((((((....)))))))).......((((....))))---- ( -40.10) >consensus AACUCCAUCUU_________________GCGACAUUUGCGAGCAAAUAAAUUCAAUUUGCUGCUGGUCAGAGGGAACUGCGACAGGUUUUUGUCGCAUUUAUCAUCGCAUCAGCAA____ ............................((((...)))).(((((((.......)))))))((((((..(((..((.((((((((....)))))))).))..).))..))))))...... (-27.53 = -26.83 + -0.69)



| Location | 12,513,003 – 12,513,102 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.56 |
| Mean single sequence MFE | -31.02 |
| Consensus MFE | -24.02 |
| Energy contribution | -24.13 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.697684 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12513003 99 - 27905053 ----UUGCUGAUGCGAUGAUAAAUGCGACAAAAACCUGUCGCAGUCCCCUCUGACCAGCAGCAAAUUGAAUUUAUUUGCUCGCAAAUGUCGC-----------------AAGAUGGAGUU ----((((....)))).......(((((((......))))))).....((((.....(((((((((.......))))))).))....(((..-----------------..))))))).. ( -29.00) >DroPse_CAF1 121 116 - 1 ----UCGCUGAUGCGAUGAUAAAUGCGACAAAAACCUGUCGCAGCUCCCUCUGACCAGCAGCAAAUUGAAUUUAUUCGCUCGCAAAUGUCGCCGACCAAUGCAUUGGCCAAGAUGGAGAC ----((((....)))).......(((((((......))))))).((((.((((((..(((((.(((.......))).))).))....)))(((((........)))))..))).)))).. ( -34.70) >DroSec_CAF1 2099 99 - 1 ----UUGCUGAUGCGAUGAUAAAUGCGACAAAAACCUGUCGCAGUCCCCUCUGACCAGCAGCAAAUUGAAUUUAUUUGCUCGCAAAUGUCGC-----------------AAGAUGGAGUU ----((((....)))).......(((((((......))))))).....((((.....(((((((((.......))))))).))....(((..-----------------..))))))).. ( -29.00) >DroSim_CAF1 2164 99 - 1 ----UUGCUGAUGCGAUGAUAAAUGCGACAAAAACCUGUCGCAGUCCCCUCUGACCAGCAGCAAAUUGAAUUUAUUUGCUCGCAAAUGUCGC-----------------AAGAUGGAGUU ----((((....)))).......(((((((......))))))).....((((.....(((((((((.......))))))).))....(((..-----------------..))))))).. ( -29.00) >DroAna_CAF1 3660 98 - 1 GCCUUUGCUGAUGCGAUGAUAAAUGCGACAAAAACCUGUCGCAGUUCCCUCCGACCAGCAGCAAAUUGAAUUUAUUUGCUCGCAAAUGUCGC-----------------CGGCC-----A (((.((((....))))........((((((.......((((.((....)).))))..(((((((((.......))))))).))...))))))-----------------.))).-----. ( -29.70) >DroPer_CAF1 3933 116 - 1 ----UCGCUGAUGCGAUGAUAAAUGCGACAAAAACCUGUCGCAGCUCCCUCUGACCAGCAGCAAAUUGAAUUUAUUCGCUCGCAAAUGUCGCCGACCAAUGCAUUGGCCAAGAUGGAGAC ----((((....)))).......(((((((......))))))).((((.((((((..(((((.(((.......))).))).))....)))(((((........)))))..))).)))).. ( -34.70) >consensus ____UUGCUGAUGCGAUGAUAAAUGCGACAAAAACCUGUCGCAGUCCCCUCUGACCAGCAGCAAAUUGAAUUUAUUUGCUCGCAAAUGUCGC_________________AAGAUGGAGUU ....((((....)))).((((..(((((((......)))))))..............(((((((((.......))))))).))...)))).............................. (-24.02 = -24.13 + 0.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:44 2006