| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,792,635 – 1,792,743 |
| Length | 108 |
| Max. P | 0.822052 |

| Location | 1,792,635 – 1,792,743 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 80.58 |
| Mean single sequence MFE | -28.98 |
| Consensus MFE | -21.04 |
| Energy contribution | -22.43 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.822052 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
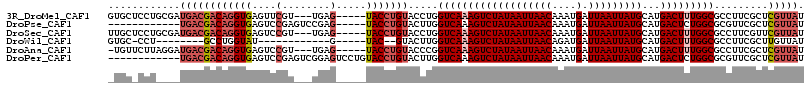
>3R_DroMel_CAF1 1792635 108 + 27905053 GUGCUCCUGCGAUGACGACAGGUGAGUUCGU---UGAG-----UACCUGUACCUGGUCAAAGUCUAUAAUUAACAAAUGAUUAAUUAUGCAUGACUUUGGCGCCUUCGCUCGUUAU .(((....)))(((((((((((((.((.(..---...)-----.))...))))))(((((((((((((((((((....).)))))))))...)))))))))........))))))) ( -31.10) >DroPse_CAF1 92871 99 + 1 ------------UGACGACAGGUGAGUCCGAGUCCGAG-----UACCUGUACUUGGUCAAAGUCUAUAAUUAACAAAUGAUUAAUUAUGCAUGACUCUGGCGCGUUCGCUCGUUAU ------------((((((...(((((..((...(((((-----((....)))))))(((.((((((((((((((....).)))))))))...)))).)))))..))))))))))). ( -27.60) >DroSec_CAF1 103373 108 + 1 UUGCUCCUGCGAUGACGACAGGUGAGUCCGU---UGAG-----UACCUGUACCUGGUCAAAGUCUAUAAUUAACAAAUGAUUAAUUAUGCAUGACUUUGGCGCCUUCGUUCGUUAU ((((....))))((((((((((((.((.(..---...)-----.))...))))))(((((((((((((((((((....).)))))))))...)))))))))........)))))). ( -31.50) >DroWil_CAF1 102230 88 + 1 GUGC-CCU--------GCCUGGUAU------------G-----UAC--GUACUUGGUCAAAGUCUAUAAUUAACAGAUGAUUAAUUAUGCAUGACUUUGGCGCCUUCGCUUGUUAU ((((-(..--------(((.(((((------------.-----...--))))).))).((((((((((((((((....).)))))))))...)))))))))))............. ( -23.10) >DroAna_CAF1 93989 107 + 1 -UGUUCUUAGGAUGACGACAGGUGAGUCCGU---UGAG-----UACCUGUACCCGGUCAAAGUCUAUAAUUAACAAAUGAUUAAUUAUGCAUGACUUUGGCGCCUUCGCUCGUUAU -.........((((((((((((((..((...---.)).-----))))))).....(((((((((((((((((((....).)))))))))...))))))))).....)).))))).. ( -29.00) >DroPer_CAF1 95455 104 + 1 ------------UGACGACAGGUGAGUCCGAGUCGGAGUCCUGUACCUGUACUUGGUCAAAGUCUAUAAUUAACAAAUGAUUAAUUAUGCAUGACUCUGGCGCGUUCGCUCGUUAU ------------((((((...(((((..((.(((((((((.((((((.......)))........(((((((((....).))))))))))).))))))))).))))))))))))). ( -31.60) >consensus _UGC_CCU____UGACGACAGGUGAGUCCGU___UGAG_____UACCUGUACCUGGUCAAAGUCUAUAAUUAACAAAUGAUUAAUUAUGCAUGACUUUGGCGCCUUCGCUCGUUAU ............((((((((((((....(........).....))))))).....(((((((((((((((((((....).)))))))))...))))))))).........))))). (-21.04 = -22.43 + 1.39)
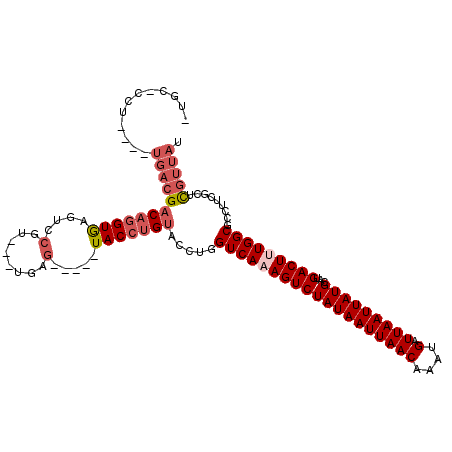
| Location | 1,792,635 – 1,792,743 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 80.58 |
| Mean single sequence MFE | -24.55 |
| Consensus MFE | -16.22 |
| Energy contribution | -17.17 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.585253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1792635 108 - 27905053 AUAACGAGCGAAGGCGCCAAAGUCAUGCAUAAUUAAUCAUUUGUUAAUUAUAGACUUUGACCAGGUACAGGUA-----CUCA---ACGAACUCACCUGUCGUCAUCGCAGGAGCAC .......((((.((((.(((((((....(((((((((.....))))))))).)))))))..(((((...(((.-----(...---..).))).))))).)))).))))........ ( -28.10) >DroPse_CAF1 92871 99 - 1 AUAACGAGCGAACGCGCCAGAGUCAUGCAUAAUUAAUCAUUUGUUAAUUAUAGACUUUGACCAAGUACAGGUA-----CUCGGACUCGGACUCACCUGUCGUCA------------ ....((((.........(((((((....(((((((((.....))))))))).))))))).((.((((....))-----)).)).))))(((......)))....------------ ( -24.60) >DroSec_CAF1 103373 108 - 1 AUAACGAACGAAGGCGCCAAAGUCAUGCAUAAUUAAUCAUUUGUUAAUUAUAGACUUUGACCAGGUACAGGUA-----CUCA---ACGGACUCACCUGUCGUCAUCGCAGGAGCAA ........(((.((((.(((((((....(((((((((.....))))))))).))))))).......((((((.-----.((.---...))...)))))))))).)))......... ( -24.40) >DroWil_CAF1 102230 88 - 1 AUAACAAGCGAAGGCGCCAAAGUCAUGCAUAAUUAAUCAUCUGUUAAUUAUAGACUUUGACCAAGUAC--GUA-----C------------AUACCAGGC--------AGG-GCAC .......((....))(((((((((....(((((((((.....))))))))).))))))(.((..(((.--...-----.------------.)))..)))--------..)-)).. ( -19.50) >DroAna_CAF1 93989 107 - 1 AUAACGAGCGAAGGCGCCAAAGUCAUGCAUAAUUAAUCAUUUGUUAAUUAUAGACUUUGACCGGGUACAGGUA-----CUCA---ACGGACUCACCUGUCGUCAUCCUAAGAACA- .......((....))..(((((((....(((((((((.....))))))))).)))))))...((((((((((.-----.((.---...))...))))))....))))........- ( -24.60) >DroPer_CAF1 95455 104 - 1 AUAACGAGCGAACGCGCCAGAGUCAUGCAUAAUUAAUCAUUUGUUAAUUAUAGACUUUGACCAAGUACAGGUACAGGACUCCGACUCGGACUCACCUGUCGUCA------------ ...(((((((....)))(((((((....(((((((((.....))))))))).)))))))........(((((....((.((((...)))).)))))))))))..------------ ( -26.10) >consensus AUAACGAGCGAAGGCGCCAAAGUCAUGCAUAAUUAAUCAUUUGUUAAUUAUAGACUUUGACCAAGUACAGGUA_____CUCA___ACGGACUCACCUGUCGUCA____AGG_GCA_ ...((((((....))..(((((((....(((((((((.....))))))))).)))))))........(((((.....................))))))))).............. (-16.22 = -17.17 + 0.95)

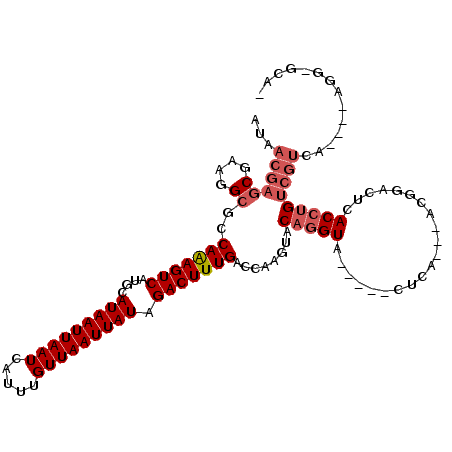
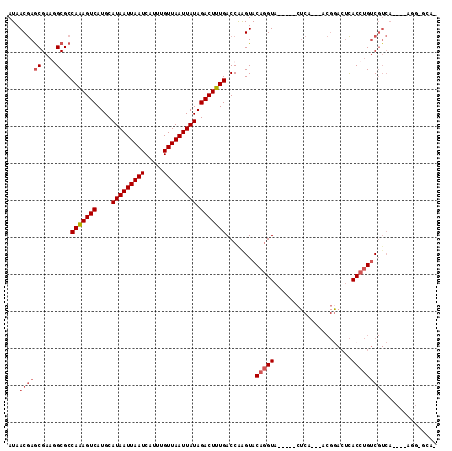
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:12 2006