
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,506,932 – 12,507,052 |
| Length | 120 |
| Max. P | 0.984002 |

| Location | 12,506,932 – 12,507,025 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.05 |
| Mean single sequence MFE | -24.39 |
| Consensus MFE | -14.53 |
| Energy contribution | -15.08 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799873 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
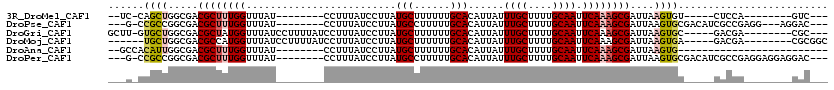
>3R_DroMel_CAF1 12506932 93 + 27905053 --UC-CAGCUGGCGACGCUUUGGUUUAU--------CCUUUAUCCUUAUGCUUUUUUGCACAUUAUUUGCUUUUGCAAUUCAAAGCGAUUAAGUGU-----CUCCA--------GUC--- --..-..(((((.(((((((......((--------(((((.......(((......)))......((((....))))...)))).))).))))))-----).)))--------)).--- ( -25.00) >DroPse_CAF1 29233 102 + 1 ---G-CCGCCGGCGACGCUUUGGUUUAU--------CCUUUAUCCUUAUGCCUUUUUGCACAUUAUUUGCUUUUGCAAUUCAAAGCGAUUAAGUGCGACAUCGCCGAGG---AGGAC--- ---.-((..((((((.((...((..((.--------....))..))...))....((((((.(((.(((((((........))))))).)))))))))..)))))).))---.....--- ( -30.30) >DroGri_CAF1 80857 103 + 1 GCUU-GUGCUGGCGACGCUAUGGUUUAUCCUUUUAUCCUUUAUCCUUAUGCUUUUUUGCACAUUAUUUGCUUUUGCAAUUCAAAGCGAUUAAGUGC-----GACGA--------CGC--- ((..-..))..(((..((...((..((.............))..))...))....((((((.(((.(((((((........))))))).)))))))-----))...--------)))--- ( -19.72) >DroMoj_CAF1 55301 101 + 1 ------UGCUGGCGACGCCAUGGUUUAUCCUUUUAUCCUUUAUCCUUAUGCUUUUUUGCACAUUAUUUGCUUUUGCAAUUCAAAGCGAUUAAGUGA-----GACGA--------CGCGGC ------.((((.((.((.(..((.....)).....(((((.((((((.(((......)))......((((....))))....))).))).))).))-----).)).--------)))))) ( -18.90) >DroAna_CAF1 26891 85 + 1 --GCCACAUUGGCGACGCUUUGGUUUAU--------CCUUUAUCCUUAUGCUUUUUUGCACAUUAUUUGCUUUUGCAAUUCAAAGCGAUUAAGUG------------------------- --(((.....)))..((((((((.....--------............(((......)))......((((....)))).))))))))........------------------------- ( -20.70) >DroPer_CAF1 30169 105 + 1 ---G-CCGCCGGCGACGCUUUGGUUUAU--------CCUUUAUCCUUAUGCCUUUUUGCACAUUAUUUGCUUUUGCAAUUCAAAGCGAUUAAGUGCGACAUCGCCGAGGAGGAGGAC--- ---(-(((.((....))...))))...(--------(((((.(((((..((..(.((((((.(((.(((((((........))))))).))))))))).)..)).))))))))))).--- ( -31.70) >consensus ___C_CCGCUGGCGACGCUUUGGUUUAU________CCUUUAUCCUUAUGCUUUUUUGCACAUUAUUUGCUUUUGCAAUUCAAAGCGAUUAAGUGC_____CACCA________GAC___ ......((((.....((((((((.........................(((......)))......((((....)))).))))))))....))))......................... (-14.53 = -15.08 + 0.56)
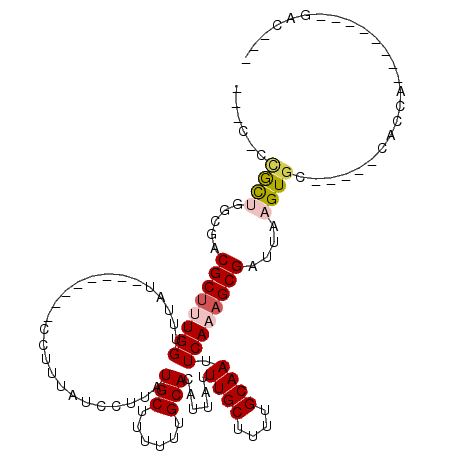

| Location | 12,506,932 – 12,507,025 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.05 |
| Mean single sequence MFE | -25.71 |
| Consensus MFE | -15.97 |
| Energy contribution | -16.17 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.984002 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12506932 93 - 27905053 ---GAC--------UGGAG-----ACACUUAAUCGCUUUGAAUUGCAAAAGCAAAUAAUGUGCAAAAAAGCAUAAGGAUAAAGG--------AUAAACCAAAGCGUCGCCAGCUG-GA-- ---..(--------(((.(-----((.(((.((((((((........)))))......(((((......)))))..)))...((--------.....)).))).))).))))...-..-- ( -23.90) >DroPse_CAF1 29233 102 - 1 ---GUCCU---CCUCGGCGAUGUCGCACUUAAUCGCUUUGAAUUGCAAAAGCAAAUAAUGUGCAAAAAGGCAUAAGGAUAAAGG--------AUAAACCAAAGCGUCGCCGGCGG-C--- ---.....---((((((((((((........(((.(((((..((((....))))....(((((......)))))....))))))--------))........)))))))))).))-.--- ( -29.69) >DroGri_CAF1 80857 103 - 1 ---GCG--------UCGUC-----GCACUUAAUCGCUUUGAAUUGCAAAAGCAAAUAAUGUGCAAAAAAGCAUAAGGAUAAAGGAUAAAAGGAUAAACCAUAGCGUCGCCAGCAC-AAGC ---((.--------.((.(-----((.(((.((((((((........)))))......(((((......)))))..))).))).......((.....))...))).))...))..-.... ( -24.20) >DroMoj_CAF1 55301 101 - 1 GCCGCG--------UCGUC-----UCACUUAAUCGCUUUGAAUUGCAAAAGCAAAUAAUGUGCAAAAAAGCAUAAGGAUAAAGGAUAAAAGGAUAAACCAUGGCGUCGCCAGCA------ ((.(((--------.((((-----((.(((.((((((((........)))))......(((((......)))))..))).))))).....((.....))..)))).)))..)).------ ( -26.80) >DroAna_CAF1 26891 85 - 1 -------------------------CACUUAAUCGCUUUGAAUUGCAAAAGCAAAUAAUGUGCAAAAAAGCAUAAGGAUAAAGG--------AUAAACCAAAGCGUCGCCAAUGUGGC-- -------------------------........(((((((..((((....))))....(((((......)))))..........--------......)))))))..(((.....)))-- ( -20.00) >DroPer_CAF1 30169 105 - 1 ---GUCCUCCUCCUCGGCGAUGUCGCACUUAAUCGCUUUGAAUUGCAAAAGCAAAUAAUGUGCAAAAAGGCAUAAGGAUAAAGG--------AUAAACCAAAGCGUCGCCGGCGG-C--- ---........((((((((((((........(((.(((((..((((....))))....(((((......)))))....))))))--------))........)))))))))).))-.--- ( -29.69) >consensus ___GCC________UGGCG_____GCACUUAAUCGCUUUGAAUUGCAAAAGCAAAUAAUGUGCAAAAAAGCAUAAGGAUAAAGG________AUAAACCAAAGCGUCGCCAGCGG_C___ .................................(((((((..((((....))))....(((((......)))))........................)))))))............... (-15.97 = -16.17 + 0.20)



| Location | 12,506,961 – 12,507,052 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 80.13 |
| Mean single sequence MFE | -23.20 |
| Consensus MFE | -12.35 |
| Energy contribution | -11.68 |
| Covariance contribution | -0.67 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12506961 91 + 27905053 UAUCCUUAUGCUUUUUUGCACAUUAUUUGCUUUUGCAAUUCAAAGCGAUUAAGUGU-----CUCCA--------GUCUCGCAACUA------GCAACUCGCAUCUCGGAU .((((..((((......((((.(((.(((((((........))))))).)))))))-----.....--------.....((.....------)).....))))...)))) ( -18.70) >DroPse_CAF1 29261 101 + 1 UAUCCUUAUGCCUUUUUGCACAUUAUUUGCUUUUGCAAUUCAAAGCGAUUAAGUGCGACAUCGCCGAGG---AGGACUCGAGAGAG------UCGCCUCGAGACUCGACG .........((..(.((((((.(((.(((((((........))))))).))))))))).)..))(((((---..(((((....)))------)).))))).......... ( -34.10) >DroSim_CAF1 29361 91 + 1 UAUCCUUAUGCUUUUUUGCACAUUAUUUGCUUUUGCAAUUCAAAGCGAUUAAGUGU-----CUCCA--------GUCUCGCAACUA------GCAACUAGCAACUCGCAU ........((((...(((((((((..(((((((........)))))))...)))))-----....(--------((......))).------))))..))))........ ( -16.20) >DroEre_CAF1 28827 97 + 1 UAUCCUUAUGCUUUUUUGCACAUUAUUUGCUUUUGCAAUUCAAAGCGAUUAAGUGU-----CUCCA--------GUCUCGCAACUCGCACUCGCAACUAGCAACUAGCAA ........((((...((((.......((((....))))......((((....((((-----....(--------((......))).)))))))).....))))..)))). ( -19.20) >DroYak_CAF1 29077 91 + 1 UAUCCUUAUGCUUUUUUGCACAUUAUUUGCUUUUGCAAUUCAAAGCGAUUAAGUGU-----CUCCA--------GUCUCGCAACUC------GCAACUAGCAACUCGCAU ........((((...((((.......((((....))))......((((...(.((.-----...))--------.).)))).....------))))..))))........ ( -16.00) >DroPer_CAF1 30197 104 + 1 UAUCCUUAUGCCUUUUUGCACAUUAUUUGCUUUUGCAAUUCAAAGCGAUUAAGUGCGACAUCGCCGAGGAGGAGGACUCGAGAGAG------UCGCCUCGAGACUCGACG .........((..(.((((((.(((.(((((((........))))))).))))))))).)..))((((((((..(((((....)))------)).))))....))))... ( -35.00) >consensus UAUCCUUAUGCUUUUUUGCACAUUAUUUGCUUUUGCAAUUCAAAGCGAUUAAGUGU_____CUCCA________GUCUCGCAACUA______GCAACUAGCAACUCGCAU .................((((.(((.(((((((........))))))).)))))))....................................((.....))......... (-12.35 = -11.68 + -0.67)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:31 2006