
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,506,056 – 12,506,163 |
| Length | 107 |
| Max. P | 0.904913 |

| Location | 12,506,056 – 12,506,163 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.38 |
| Mean single sequence MFE | -33.73 |
| Consensus MFE | -19.19 |
| Energy contribution | -19.58 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
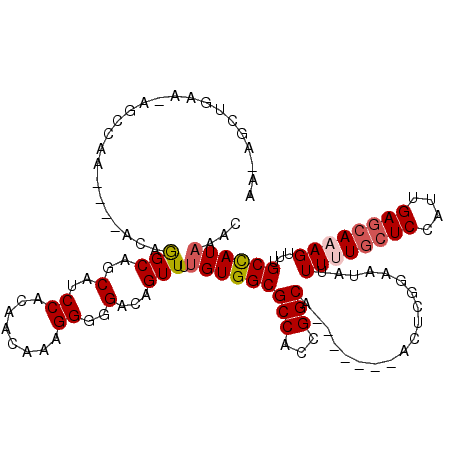
>3R_DroMel_CAF1 12506056 107 + 27905053 UA-AGCUGAA-AGCCAA----ACAGGCAGCUUCCCCAACAAAGGGGGACAGUUUUGUGGCGCCACCGGCA-------ACUCGGAAUAUUUUUGCUCCAUUGAGCAAAGUUUGCCAUAAAC ..-(((((..-.(((..----...)))...((((((......)))))))))))(((((((....((((..-------..)))).....((((((((....))))))))...))))))).. ( -38.60) >DroPse_CAF1 28455 100 + 1 C--------------------AGCGGCAGCAUCCACAACAAUGGGGGACAGUUAUGUGGCGCCACAGGCGGGAGGAGACUUCGAAUAUUUGUGCUCCAUUGAGCAAAGUUUGCUAUAAAC .--------------------.((....))........((((((((.((((.(((....((((...))))((((....))))..))).)))).))))))))((((.....))))...... ( -33.30) >DroSec_CAF1 27523 107 + 1 UA-AGCUGAA-AGCCAA----ACAGGCAGCUUCCCCAACAAAGGGGGACAGUUUUGUGGCGCCACCGGCA-------ACUCGGAAUAUUUUUUCUCCAUUGAGCAAAGUUUGCCAUAAAC ..-(((((..-.(((..----...)))...((((((......)))))))))))((((((((((...))).-------.(((((((......)))).....)))........))))))).. ( -32.30) >DroYak_CAF1 28175 107 + 1 AA-AGCUGUA-AGCCAA----ACAGGCAGCUUCCCCAACAAAGGGGGACAGUUUUGUGGCGCCACCGGCA-------ACUCGGAAUAUUUUUGCUCCAUUGAGCAAAGUUUGCCAUAAAC ((-((((((.-.(((..----...)))....(((((......)))))))))))))(((((....((((..-------..)))).....((((((((....))))))))...))))).... ( -39.00) >DroAna_CAF1 26202 113 + 1 AACAGCCAAAGAGCCAGCAGCCCAAGCAGCAUCCACAACAAUGGGGGACAGUUUUGUGGCGCCACGGGCG-------AGCCGGAAUAUUUUUGCUCCAUUGAACAAAGUUUGCCAUCAAC ...(((.(((((.((.((.((((.....((..((((((.(((.(....).))))))))).))...)))).-------.)).))....))))))))......................... ( -27.50) >DroPer_CAF1 29387 100 + 1 C--------------------AACAGCAGCAUCCACAACAAUGGGGGACAGUUAUGUGGCGCCACAGGCGGGAGGAGACUUCGAAUAUUUGUGCUCCAUUGAGCAAAGUUUGCUAUAAAC .--------------------......((((..(....((((((((.((((.(((....((((...))))((((....))))..))).)))).))))))))......)..))))...... ( -31.70) >consensus AA_AGCUGAA_AGCCAA____ACAGGCAGCAUCCACAACAAAGGGGGACAGUUUUGUGGCGCCACCGGCA_______ACUCGGAAUAUUUUUGCUCCAUUGAGCAAAGUUUGCCAUAAAC ........................(((..(..((........))..)...))).(((((((((...)))...................((((((((....))))))))...))))))... (-19.19 = -19.58 + 0.39)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:28 2006