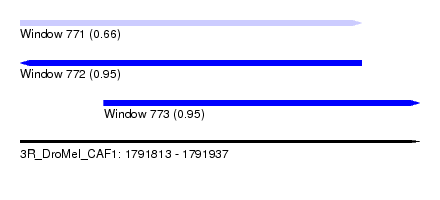
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,791,813 – 1,791,937 |
| Length | 124 |
| Max. P | 0.951861 |

| Location | 1,791,813 – 1,791,919 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 82.90 |
| Mean single sequence MFE | -18.45 |
| Consensus MFE | -16.70 |
| Energy contribution | -16.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.663427 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1791813 106 + 27905053 --GCAU-CAGGAGAUGAAAA----AACGAAAACAAAAUAUUAUGUAAUUUUACAUUCAAGUGAGCGGAAAAUAUAAAUAAGCAGCAGGUGCUCAUUUCCUGUUCCGCAGCAAA --((..-((((((((((...----.............((((.((((.((((...(((....)))..)))).)))))))).(((.....)))))))))))))....))...... ( -20.20) >DroVir_CAF1 119854 102 + 1 AAGAAUA-------AAUAAA----AAUAAAGAGAAAAUAUCAUGUAAUUUUACAUUCAAGUGAGCGGAAAAUAUAAAUAAGCAGCAGGUGCUCAUUUCCUGUUCCGCAGCAAA ..((((.-------......----......((.......))..(((....)))))))..((..((((((.....((((.((((.....)))).))))....)))))).))... ( -16.80) >DroGri_CAF1 105488 90 + 1 AAGA-----------------------ACCGAGAAUAUAACAUGUAAUUUUACAUUCAAGUGAGCGGAAAAUAUAAAUAAGCAGCAGGUGCUCAUUUCCUGUUCCGCAGCAAA ....-----------------------...............(((....((((......))))((((((.....((((.((((.....)))).))))....)))))).))).. ( -16.70) >DroWil_CAF1 101605 102 + 1 --GAAU-AAGAAG---CGAA----AACGAAAA-AAAAUAUUAUGUAAUUUUACAUUCAAGUGAGCGGAAAAUAUAAAUAAGCAGCAGGUGCUCAUUUCCUGUUCCGCAGCAAA --....-.....(---(...----........-........(((((....)))))........((((((.....((((.((((.....)))).))))....)))))).))... ( -18.30) >DroMoj_CAF1 111958 109 + 1 ACGAAUACAAGAAAUAUAUA----UAUAAAGAGAAAAUAUCAUGUAAUUUUACAUUCAAGUGAGCGGAAAAUAUAAAUAAGCAGCAGGUGCUCAUUUCCUGUUCCGCAGCAAA ..((((...((((.((((((----(((.........)))).))))).))))..))))..((..((((((.....((((.((((.....)))).))))....)))))).))... ( -18.90) >DroAna_CAF1 93275 110 + 1 --GCAU-CAGGAGCUGAAAAAAAAAAUGAAAACAAAAUAUUAUGUAAUUUUACAUUCAAGUGAGCGGAAAAUAUAAAUAAGCAGCAGGUGCUCAUUUCCUGUUCCGCAGCAAA --((.(-(((...))))...............((.......(((((....))))).....)).((((((.....((((.((((.....)))).))))....)))))).))... ( -19.80) >consensus __GAAU_CAGGAG_U_AAAA____AAUAAAAAGAAAAUAUCAUGUAAUUUUACAUUCAAGUGAGCGGAAAAUAUAAAUAAGCAGCAGGUGCUCAUUUCCUGUUCCGCAGCAAA ..........................................(((....((((......))))((((((.....((((.((((.....)))).))))....)))))).))).. (-16.70 = -16.70 + -0.00)

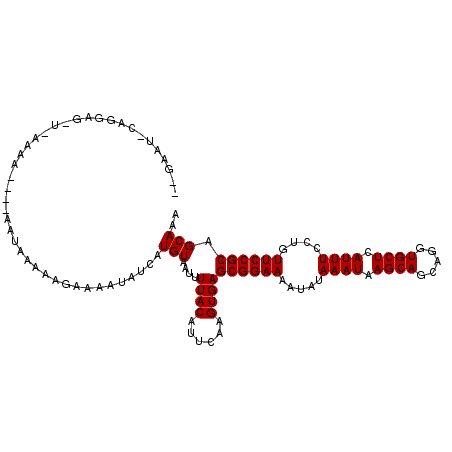
| Location | 1,791,813 – 1,791,919 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 82.90 |
| Mean single sequence MFE | -22.44 |
| Consensus MFE | -19.70 |
| Energy contribution | -19.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951169 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1791813 106 - 27905053 UUUGCUGCGGAACAGGAAAUGAGCACCUGCUGCUUAUUUAUAUUUUCCGCUCACUUGAAUGUAAAAUUACAUAAUAUUUUGUUUUCGUU----UUUUCAUCUCCUG-AUGC-- ......((((((.(..(((((((((.....)))))))))...).))))))......(((..((((((........))))))..)))...----....((((....)-))).-- ( -24.20) >DroVir_CAF1 119854 102 - 1 UUUGCUGCGGAACAGGAAAUGAGCACCUGCUGCUUAUUUAUAUUUUCCGCUCACUUGAAUGUAAAAUUACAUGAUAUUUUCUCUUUAUU----UUUAUU-------UAUUCUU ......((((((.(..(((((((((.....)))))))))...).))))))......(((((.(((((.....((.....)).....)))----))....-------))))).. ( -21.00) >DroGri_CAF1 105488 90 - 1 UUUGCUGCGGAACAGGAAAUGAGCACCUGCUGCUUAUUUAUAUUUUCCGCUCACUUGAAUGUAAAAUUACAUGUUAUAUUCUCGGU-----------------------UCUU ......((((((.(..(((((((((.....)))))))))...).))))))..(((.((((((((.........))))))))..)))-----------------------.... ( -22.40) >DroWil_CAF1 101605 102 - 1 UUUGCUGCGGAACAGGAAAUGAGCACCUGCUGCUUAUUUAUAUUUUCCGCUCACUUGAAUGUAAAAUUACAUAAUAUUUU-UUUUCGUU----UUCG---CUUCUU-AUUC-- ...((.((((((.(..(((((((((.....)))))))))...).))))))........(((((....)))))........-........----...)---).....-....-- ( -21.20) >DroMoj_CAF1 111958 109 - 1 UUUGCUGCGGAACAGGAAAUGAGCACCUGCUGCUUAUUUAUAUUUUCCGCUCACUUGAAUGUAAAAUUACAUGAUAUUUUCUCUUUAUA----UAUAUAUUUCUUGUAUUCGU (((((.((((((.(..(((((((((.....)))))))))...).))))))((....))..)))))..((((.(((((............----...)))))...))))..... ( -20.86) >DroAna_CAF1 93275 110 - 1 UUUGCUGCGGAACAGGAAAUGAGCACCUGCUGCUUAUUUAUAUUUUCCGCUCACUUGAAUGUAAAAUUACAUAAUAUUUUGUUUUCAUUUUUUUUUUCAGCUCCUG-AUGC-- ...(((((((((.(..(((((((((.....)))))))))...).)))))).....((((..((((((........))))))..))))...........))).....-....-- ( -25.00) >consensus UUUGCUGCGGAACAGGAAAUGAGCACCUGCUGCUUAUUUAUAUUUUCCGCUCACUUGAAUGUAAAAUUACAUAAUAUUUUCUCUUCAUU____UUUA_A_CUCCUG_AUUC__ ......((((((.(..(((((((((.....)))))))))...).))))))........(((((....)))))......................................... (-19.70 = -19.70 + -0.00)


| Location | 1,791,839 – 1,791,937 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.69 |
| Mean single sequence MFE | -23.10 |
| Consensus MFE | -21.43 |
| Energy contribution | -21.52 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.951861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1791839 98 + 27905053 AAAAUAUUAUGUAAUUUUACAUUCAAGUGAGCGGAAAAUAUAAAUAAGCAGCAGGUGCUCAUUUCCUGUUCCGCAGCAAACAUGGUACAGUCA--------GCAUA-------------- ....((((((((....((((......))))((((((.....((((.((((.....)))).))))....)))))).....))))))))......--------.....-------------- ( -21.40) >DroVir_CAF1 119876 97 + 1 AAAAUAUCAUGUAAUUUUACAUUCAAGUGAGCGGAAAAUAUAAAUAAGCAGCAGGUGCUCAUUUCCUGUUCCGCAGCAAACAUGGUACACUC---------GCACA-------------- ....((((((((....((((......))))((((((.....((((.((((.....)))).))))....)))))).....)))))))).....---------.....-------------- ( -23.50) >DroPse_CAF1 92391 106 + 1 AAAAUAUUAUGUAAUUUUACAUUCAAGUGAGCGGAAAAUAUAAAUAAGCAGCAGGUGCUCAUUUCCUGUUCCGCAGCAAACAUGGUACAGCCAGCCAGCGAGCACA-------------- ........(((((....)))))....(((.((((((.....((((.((((.....)))).))))....)))))).((.....(((.....)))....))...))).-------------- ( -23.40) >DroGri_CAF1 105498 97 + 1 AAUAUAACAUGUAAUUUUACAUUCAAGUGAGCGGAAAAUAUAAAUAAGCAGCAGGUGCUCAUUUCCUGUUCCGCAGCAAACAUGCUACAGUA---------GCACA-------------- ........(((((....)))))....(((.((((((.....((((.((((.....)))).))))....))))))((((....))))......---------.))).-------------- ( -21.50) >DroMoj_CAF1 111987 111 + 1 AAAAUAUCAUGUAAUUUUACAUUCAAGUGAGCGGAAAAUAUAAAUAAGCAGCAGGUGCUCAUUUCCUGUUCCGCAGCAAACAUGGCACAGUA---------GCACAAGUACUCUGGCACA .......(((((....((((......))))((((((.....((((.((((.....)))).))))....)))))).....)))))((.(((.(---------(........)))))))... ( -25.40) >DroPer_CAF1 94963 106 + 1 AAAAUAUUAUGUAAUUUUACAUUCAAGUGAGCGGAAAAUAUAAAUAAGCAGCAGGUGCUCAUUUCCUGUUCCGCAGCAAACAUGGUACAGCCAGCCAGCGAGCACA-------------- ........(((((....)))))....(((.((((((.....((((.((((.....)))).))))....)))))).((.....(((.....)))....))...))).-------------- ( -23.40) >consensus AAAAUAUCAUGUAAUUUUACAUUCAAGUGAGCGGAAAAUAUAAAUAAGCAGCAGGUGCUCAUUUCCUGUUCCGCAGCAAACAUGGUACAGUC_________GCACA______________ ....((((((((....((((......))))((((((.....((((.((((.....)))).))))....)))))).....))))))))................................. (-21.43 = -21.52 + 0.08)

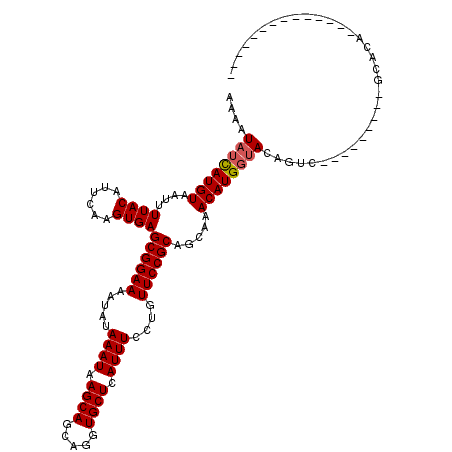
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:10 2006