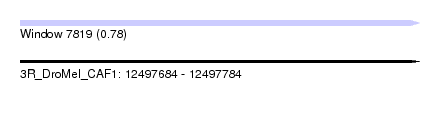
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,497,684 – 12,497,784 |
| Length | 100 |
| Max. P | 0.778898 |

| Location | 12,497,684 – 12,497,784 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 78.10 |
| Mean single sequence MFE | -19.95 |
| Consensus MFE | -14.61 |
| Energy contribution | -14.70 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.778898 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12497684 100 + 27905053 UAAUAUUAAAUGUUACACCAGGGACACAUUUCGAUGAAAUAUUUCGUAAAAAGUGUCUACGAGAGAAAAAUUUGCUCUCUUUCUGC------U----CCU-UUCGCCGCCC----- ..................(((((((((.(((..(((((....)))))..)))))))))..(((((.........)))))...))).------.----...-..........----- ( -18.30) >DroPse_CAF1 20366 112 + 1 UAAUAUUAAAUGUUACACCAGGGACACAUUUCGAUGAAAUAUUUCGUAAAAAGUGUCUACGAGAGAAAAAUUUGCUCCCGCUCUUUGCGGGCC---AACG-CCAGAGUCUCAGCUU ...........(((.......((((((.(((..(((((....)))))..)))))))))....((((....(((((.(((((.....)))))..---...)-.)))).))))))).. ( -26.10) >DroYak_CAF1 19417 104 + 1 UAAUAUUAAAUGUUACACCAGGGACACAUUUCGAUGAAAUAUUUCGUAAAAAGUGUCUACGAGAGAAAAAUUUGCUCUCUUUCUCC------C---ACCU-CCCACCUCCCACC-- ...................((((((((.(((..(((((....)))))..))))))))...(((((.........))))).......------.---.)))-.............-- ( -17.60) >DroMoj_CAF1 44788 97 + 1 UAAUAUUAAAUGUUACACCAGGGACACAUUUCGAUGAAAUAUUUCGUAAAAAGUGUCUACGAGCGAAAAAUGCGAGCCUGCGCCGC------U----CCUAUUCUCA--------- .....................((((((.(((..(((((....)))))..)))))))))..(((((......(((......))))))------)----).........--------- ( -19.90) >DroAna_CAF1 18141 103 + 1 UAAUAUUAAAUGUUACACCAGGGACACAUUUCGAUGAAAUAUUUCGUAAAAAGUGUCUACGAGAGAAAAAUUUGCUCUCGCCCCCA------UUCUUCAU-GCCGCUUUC------ ....................(((((((.(((..(((((....)))))..))))))))..((((((.........))))))...)).------........-.........------ ( -19.10) >DroPer_CAF1 21087 89 + 1 UAAUAUUAAAUGUUACACCAGGGACACAUUUCGAUGAAAUAUUUCGUAAAAAGUGUCUACGAGAGAAAAAUUUGCUCUCUCUC---------------------------CAGCUU ....................(((((((.(((..(((((....)))))..))))))))...((((((.........)))))).)---------------------------)..... ( -18.70) >consensus UAAUAUUAAAUGUUACACCAGGGACACAUUUCGAUGAAAUAUUUCGUAAAAAGUGUCUACGAGAGAAAAAUUUGCUCUCGCUCUCC______U____CCU_CCCGCCUCCC_____ .....................((((((.(((..(((((....)))))..)))))))))..(((((.........)))))..................................... (-14.61 = -14.70 + 0.09)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:22 2006