
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,490,763 – 12,490,894 |
| Length | 131 |
| Max. P | 0.988290 |

| Location | 12,490,763 – 12,490,858 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 85.29 |
| Mean single sequence MFE | -34.27 |
| Consensus MFE | -22.51 |
| Energy contribution | -22.60 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.616767 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12490763 95 - 27905053 UUUUAU------UGGCCCGUUUUUCGGGUGAGUAGCGGAAAUGUUGCACAUUUCCGUUCCACAAGCAGCGCCAUAACUCCCAUCGAUAAUUAUCGGGGACU ......------(((((((.....)))(((.(.((((((((((.....)))))))))))))).......))))....((((..((((....)))))))).. ( -32.50) >DroPse_CAF1 14254 97 - 1 UUUUACGAGUUGUGGCCCGUUUU-UGGGUAAAGUGCGGAAAUGUUGCACAUUUCCGUUCCCGA-GC--UGCCAUAACUCCAAUCGAUAAUUAUCGGGGGCA ......((((((((((..((..(-((((......(((((((((.....))))))))).)))))-))--.))))))))))(..(((((....)))))..).. ( -36.80) >DroEre_CAF1 12500 95 - 1 UUUUAU------UGGCCCGUUUUUCGGGUGAGCAGCGGAAAUGUUGCACAUUUCCGUUCCAGAAGCAGCGCCAUAACUCCCAUCGAUAAUUAUCGGGGACU ......------(((((.((((((.(((......(((((((((.....)))))))))))))))))).).))))....((((..((((....)))))))).. ( -30.80) >DroYak_CAF1 12429 95 - 1 UUUUAU------UGGCCCGUUUUUCGGGUGAGUAGCGGAAAUGUUGCACAUUUCCGUUCCAGAAGCAGCGCCAUAACUCCCAUCGAUAAUUAUCGGCGACU ...(((------(.(((((.....))))).))))(((((((((.....))))))))).........((((((((((.((.....))...)))).)))).)) ( -29.20) >DroAna_CAF1 11900 93 - 1 UUUUAU------UGGGCCGUUUUUCGGGUGAAGAGCGGAAAUGUUGCACAUUUCCGUUCCCGAAUCGG--CCAUAACUUCCAUCGAUAAUUAUCGGGGACG ......------..(((((...((((((.....((((((((((.....)))))))))))))))).)))--))......(((.(((((....)))))))).. ( -38.90) >DroPer_CAF1 14782 96 - 1 UUUUACGAGUUGUGGCCCGUU-U-UGGGUAAAGUGCGGAAAUGUUGCACAUUUCCGUUCCCGA-GC--UGCCAUAACUCCAAUCGAUAAUUAUCGGGGGCA ......((((((((((..(((-(-.(((......(((((((((.....))))))))).)))))-))--.))))))))))(..(((((....)))))..).. ( -37.40) >consensus UUUUAU______UGGCCCGUUUUUCGGGUGAAGAGCGGAAAUGUUGCACAUUUCCGUUCCAGAAGCAGCGCCAUAACUCCCAUCGAUAAUUAUCGGGGACU ............(((((((.....))).......(((((((((.....)))))))))............))))....((((..((((....)))))))).. (-22.51 = -22.60 + 0.09)



| Location | 12,490,789 – 12,490,894 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.26 |
| Mean single sequence MFE | -30.33 |
| Consensus MFE | -13.05 |
| Energy contribution | -14.22 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.43 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12490789 105 + 27905053 UAUGGCGCUGCUUGUG-GAACGGAAAUGUGCAACAUUUCCGCUACUCACCCGAAAAACG----GGCCA------AUAAAAAUCAGCGCCAAGUGUCAAAUCUAAAUAAUGACUCCC---- ..((((((((.((.((-(..((((((((.....)))))))).......((((.....))----)))))------.....)).)))))))).(.((((...........)))).)..---- ( -35.50) >DroPse_CAF1 14280 110 + 1 UAUGGCA--GC-UCGG-GAACGGAAAUGUGCAACAUUUCCGCACUUUACCCA-AAAACG----GGCCACAACUCGUAAAAAUCAGCGCCAAAUGUCAAAUCUAACUACCGAUAGCC-GGC ..((((.--((-(.((-(((((((((((.....))))))))....)).))).-...(((----((......))))).......))))))).................(((.....)-)). ( -29.10) >DroGri_CAF1 63341 107 + 1 UAUGGAGCAGCAACUAGAAACGGAAAUGUGCAACAUUUCCGCAGGCUCUUC--AAAUCGUGUUGCCCA------AUAAAAAUCAACGC-AAGUGUCAAACAUAAAUUACACUGCGC---- ..(((.((((((....((..((((((((.....))))))))..((....))--...)).)))))))))------...........(((-(.((((............)))))))).---- ( -28.00) >DroEre_CAF1 12526 105 + 1 UAUGGCGCUGCUUCUG-GAACGGAAAUGUGCAACAUUUCCGCUGCUCACCCGAAAAACG----GGCCA------AUAAAAAUCAGCGCCAAGUGUCAAAUCUAAAUAAUGAUUCCC---- ..((((((((....((-(..((((((((.....)))))))).......((((.....))----)))))------........)))))))).......((((........))))...---- ( -32.50) >DroAna_CAF1 11926 107 + 1 UAUGG--CCGAUUCGG-GAACGGAAAUGUGCAACAUUUCCGCUCUUCACCCGAAAAACG----GCCCA------AUAAAAAUCAACGCCAAGUGUCAAAUCUAAAUAGCGAUUCCCCGUC ...((--(((.(((((-(..((((((((.....)))))))).......))))))...))----)))..------...........(((...................))).......... ( -27.81) >DroPer_CAF1 14808 109 + 1 UAUGGCA--GC-UCGG-GAACGGAAAUGUGCAACAUUUCCGCACUUUACCCA-A-AACG----GGCCACAACUCGUAAAAAUCAGCGCCAAAUGUCAAAUCUAACUACCGAUAGCC-GGC ..((((.--((-(.((-(((((((((((.....))))))))....)).))).-.-.(((----((......))))).......))))))).................(((.....)-)). ( -29.10) >consensus UAUGGCGC_GCUUCGG_GAACGGAAAUGUGCAACAUUUCCGCACUUCACCCG_AAAACG____GGCCA______AUAAAAAUCAGCGCCAAGUGUCAAAUCUAAAUAACGAUUCCC____ ..((((((............((((((((.....))))))))......................(((....................)))..))))))....................... (-13.05 = -14.22 + 1.17)

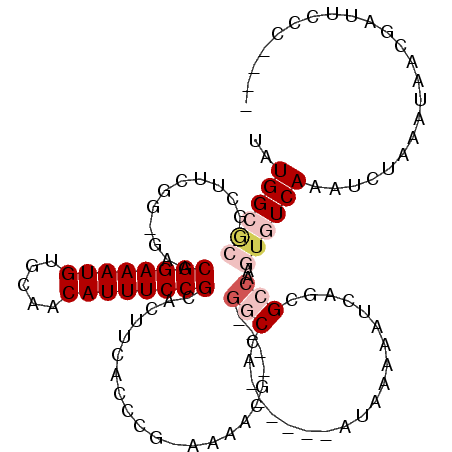

| Location | 12,490,789 – 12,490,894 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.26 |
| Mean single sequence MFE | -36.73 |
| Consensus MFE | -15.13 |
| Energy contribution | -15.77 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.41 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.945560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12490789 105 - 27905053 ----GGGAGUCAUUAUUUAGAUUUGACACUUGGCGCUGAUUUUUAU------UGGCC----CGUUUUUCGGGUGAGUAGCGGAAAUGUUGCACAUUUCCGUUC-CACAAGCAGCGCCAUA ----....((((...........))))...((((((((.....(((------(.(((----((.....))))).))))(((((((((.....)))))))))..-......)))))))).. ( -42.30) >DroPse_CAF1 14280 110 - 1 GCC-GGCUAUCGGUAGUUAGAUUUGACAUUUGGCGCUGAUUUUUACGAGUUGUGGCC----CGUUUU-UGGGUAAAGUGCGGAAAUGUUGCACAUUUCCGUUC-CCGA-GC--UGCCAUA (((-(...((((((.(((((((.....))))))))))))).....)).))((((((.----.((..(-((((......(((((((((.....))))))))).)-))))-))--.)))))) ( -35.00) >DroGri_CAF1 63341 107 - 1 ----GCGCAGUGUAAUUUAUGUUUGACACUU-GCGUUGAUUUUUAU------UGGGCAACACGAUUU--GAAGAGCCUGCGGAAAUGUUGCACAUUUCCGUUUCUAGUUGCUGCUCCAUA ----(((((((((............)))).)-))))..........------((((((...(((((.--(((......(((((((((.....)))))))))))).))))).))).))).. ( -29.40) >DroEre_CAF1 12526 105 - 1 ----GGGAAUCAUUAUUUAGAUUUGACACUUGGCGCUGAUUUUUAU------UGGCC----CGUUUUUCGGGUGAGCAGCGGAAAUGUUGCACAUUUCCGUUC-CAGAAGCAGCGCCAUA ----..(((((........)))))......((((((((.(((((..------(.(((----((.....))))).)(.((((((((((.....)))))))))).-)))))))))))))).. ( -40.90) >DroAna_CAF1 11926 107 - 1 GACGGGGAAUCGCUAUUUAGAUUUGACACUUGGCGUUGAUUUUUAU------UGGGC----CGUUUUUCGGGUGAAGAGCGGAAAUGUUGCACAUUUCCGUUC-CCGAAUCGG--CCAUA ...(..(((((........)))))..)...................------..(((----((...((((((.....((((((((((.....)))))))))))-))))).)))--))... ( -37.20) >DroPer_CAF1 14808 109 - 1 GCC-GGCUAUCGGUAGUUAGAUUUGACAUUUGGCGCUGAUUUUUACGAGUUGUGGCC----CGUU-U-UGGGUAAAGUGCGGAAAUGUUGCACAUUUCCGUUC-CCGA-GC--UGCCAUA (((-(...((((((.(((((((.....))))))))))))).....)).))((((((.----.(((-(-.(((......(((((((((.....))))))))).)-))))-))--.)))))) ( -35.60) >consensus ____GGGAAUCGUUAUUUAGAUUUGACACUUGGCGCUGAUUUUUAU______UGGCC____CGUUUU_CGGGUAAAGAGCGGAAAUGUUGCACAUUUCCGUUC_CAGAAGC_GCGCCAUA ..............................((((((.(...(((((.......(((......)))......)))))..(((((((((.....))))))))).........).)))))).. (-15.13 = -15.77 + 0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:15 2006