
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,483,315 – 12,483,411 |
| Length | 96 |
| Max. P | 0.552285 |

| Location | 12,483,315 – 12,483,411 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 79.42 |
| Mean single sequence MFE | -19.08 |
| Consensus MFE | -13.04 |
| Energy contribution | -13.37 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.552285 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12483315 96 + 27905053 GAGCGUACAACUCACGAUCAAUUAACGGCUCAAAACGAGAUCAAAAGCAAUUUCGCAUGCAAGCGGGU----------AGCCCGCGAAG---ACUCUCUCAAACAACUC ((((.......................)))).....((((((....(((........)))..(((((.----------..)))))...)---).))))........... ( -19.90) >DroSec_CAF1 4820 96 + 1 GAGCGUUCAACUCACGAUCAAUUAACGGAUCGAAACGAGAUCAAAAGCAAUUUCGCAUGCAAGCGGGC----------AGUCCGCGAAC---UCUUUCUCAAACAACUC (((...........(((((........)))))....(((.((....(((........)))..((((..----------...)))))).)---))...)))......... ( -21.20) >DroSim_CAF1 3640 96 + 1 GAGCGUACAACUCACGAUCAAUUAACGGAUCGAAACGAGAUCAAAAGCAAUUUCGCAUGCAAGCGGGC----------AGCCCGCGAAC---UCUUUCUCAAACAACUC (((...........(((((........)))))....(((.((....(((........)))..(((((.----------..))))))).)---))...)))......... ( -23.20) >DroYak_CAF1 4871 89 + 1 GAGCGUACAACUCACGAUCAAUUAACGG-----AACGAGAUCAAAAGCAAUUUCGCAUGCAAGCGGGC----------AGCCCGCGAA-----CUCUCUCAAACAACUG ((.(((.......))).))........(-----(..(((.((....(((........)))..(((((.----------..))))))).-----)))..))......... ( -18.10) >DroAna_CAF1 4568 88 + 1 GAUCGUACAACUUACGAUCAAUCAACGG----AAACGAGAUCAAAAGCAAUUUCGCAUGCAUUCUC--------------UCUAAAAAC---AAACUCGCAAACAACUC (((((((.....))))))).......(.----...)((((......((......))......))))--------------.........---................. ( -15.10) >DroPer_CAF1 6506 104 + 1 UAGCAUACAACUUACGAUCAAUUAACGG-----AACGAGAUCAAAAGCAAUUUCGCAUGCGAGCGGGAAAACAAAGCGAGCUCGCGAACUACUAUUUCUCAAAUAUUCU ............................-----...((((......((......)).(((((((...............)))))))..........))))......... ( -16.96) >consensus GAGCGUACAACUCACGAUCAAUUAACGG____AAACGAGAUCAAAAGCAAUUUCGCAUGCAAGCGGGC__________AGCCCGCGAAC___UCUCUCUCAAACAACUC ..((((.........((((......((........)).))))....((......))))))..((((((...........))))))........................ (-13.04 = -13.37 + 0.32)

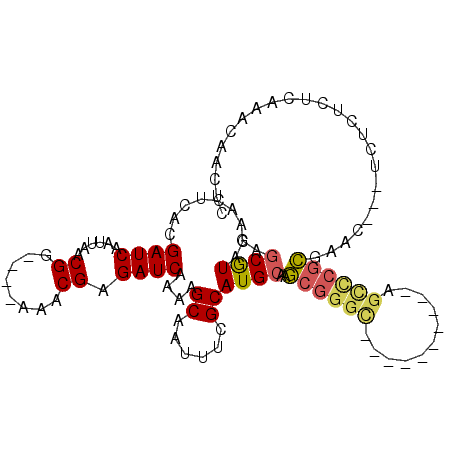
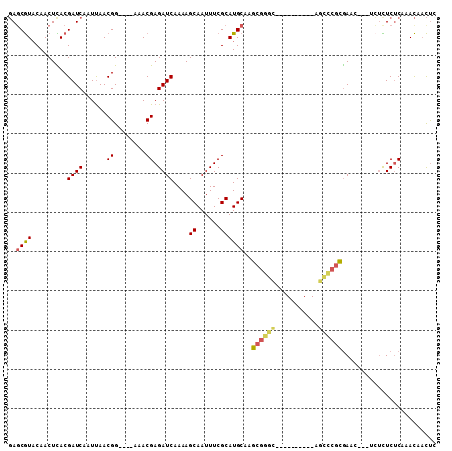
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:07 2006