| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,421,262 – 12,421,421 |
| Length | 159 |
| Max. P | 0.575191 |

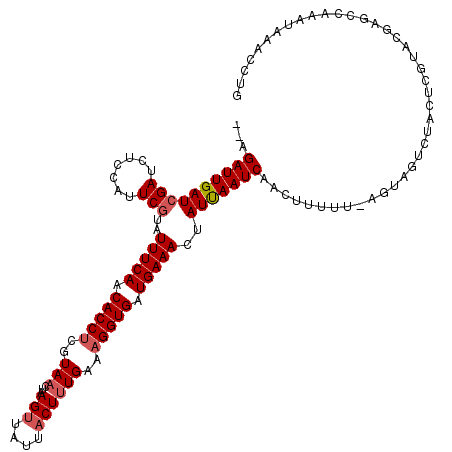
| Location | 12,421,262 – 12,421,382 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.99 |
| Mean single sequence MFE | -20.15 |
| Consensus MFE | -13.62 |
| Energy contribution | -14.36 |
| Covariance contribution | 0.74 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.505317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12421262 120 + 27905053 GAAGUAUGCUCAUAAAAAAAAAGGAAAAUCGUAAACUACAAUAGAUUGAUCGAUCACCAUUCGUAUUUCAACACCUCGUAACUAAGUUAUUACUUUGAAAGGUGAUGAAACUAUUAAUCA (.(((.(((.....................))).))).)....((((((((((.......)))..(((((.(((((..(((...(((....))))))..))))).)))))..))))))). ( -17.70) >DroSec_CAF1 20157 114 + 1 GAAGUAUGCUCAUAAAAAUAAA-UAAAAUCGUAAACU-----AGAUUGAUCGAUCUCCAUUCGUAUUUCAACACCUCGUAACUAAGUUAUUACUUUGAAAGGUGAUGAAACUAUUAAUCA ..(((.(((.............-.......))).)))-----.((((((((((.......)))..(((((.(((((..(((...(((....))))))..))))).)))))..))))))). ( -17.65) >DroSim_CAF1 19904 104 + 1 GAAG----CUCAUAAAA-------AAAAUCGUAAACU-----AGAUUGAUCGAUCUCCAUUCGUAUUUCAACACCCCGUAACUAAGUUAUUACUUUGAAAGGUGAUGAAACUAUUAAUCA ....----.........-------.............-----.((((((((((.......)))..(((((.((((........((((....)))).....)))).)))))..))))))). ( -14.82) >DroEre_CAF1 20498 102 + 1 AAAGUAUGCUAAUAAGCGCAGU-UAAAAGCGUAAACU-----AGAUUGAUA-----------GUAUUUCAACACCUCGUAACUAAGUUAUUACU-UGAAAGGUGAUGAAACUAUCAAUCA ...............((((...-.....)))).....-----.((((((((-----------((..((((.(((((..(((.(((....))).)-))..))))).)))))))))))))). ( -29.20) >DroYak_CAF1 20840 114 + 1 AAGGUAUGCUAAUAAGCGAAGC-UAAAAGCGUAAACU-----AGAUUGAUCGAUCUCCAUUCGCAUUUCAACACCUCGUAACUAAGCCAUUACUUUGAAAGGUGAUGAAACAAUUAAUCA ..(((..........(((((((-.....)).......-----(((((....)))))...))))).(((((.(((((..(((.(((....)))..)))..))))).)))))......))). ( -21.40) >consensus GAAGUAUGCUCAUAAAAAAAAA_UAAAAUCGUAAACU_____AGAUUGAUCGAUCUCCAUUCGUAUUUCAACACCUCGUAACUAAGUUAUUACUUUGAAAGGUGAUGAAACUAUUAAUCA ...........................................((((((((((.......)))..(((((.(((((..(((...(((....))))))..))))).)))))..))))))). (-13.62 = -14.36 + 0.74)

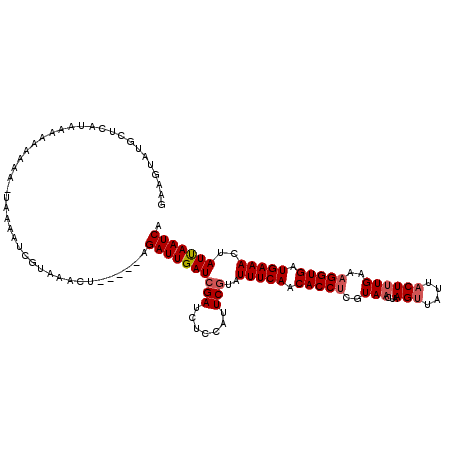
| Location | 12,421,302 – 12,421,421 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.73 |
| Mean single sequence MFE | -24.76 |
| Consensus MFE | -13.62 |
| Energy contribution | -14.36 |
| Covariance contribution | 0.74 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575191 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12421302 119 + 27905053 AUAGAUUGAUCGAUCACCAUUCGUAUUUCAACACCUCGUAACUAAGUUAUUACUUUGAAAGGUGAUGAAACUAUUAAUCAACUUUUU-AGUAGUGUACUCGUACGAGCCAAAUAAACCUG ...((((....))))....(((((((((((.(((((..(((...(((....))))))..))))).))))((((((((........))-))))))......)))))))............. ( -24.60) >DroSec_CAF1 20193 117 + 1 --AGAUUGAUCGAUCUCCAUUCGUAUUUCAACACCUCGUAACUAAGUUAUUACUUUGAAAGGUGAUGAAACUAUUAAUCACCUUUUU-AGUAGUCUACUCGUACGAGCCAAAUAAACCUG --...((((.(((.......)))....))))...((((((....(((.((((((..((((((((((..........)))))))))).-))))))..)))..))))))............. ( -28.50) >DroSim_CAF1 19930 117 + 1 --AGAUUGAUCGAUCUCCAUUCGUAUUUCAACACCCCGUAACUAAGUUAUUACUUUGAAAGGUGAUGAAACUAUUAAUCACCUUUUU-AGUAGUCUACUCGUACGAGCCAAAUAAACCUG --...((((.(((.......)))....)))).....((((....(((.((((((..((((((((((..........)))))))))).-))))))..)))..))))............... ( -24.70) >DroEre_CAF1 20534 98 + 1 --AGAUUGAUA-----------GUAUUUCAACACCUCGUAACUAAGUUAUUACU-UGAAAGGUGAUGAAACUAUCAAUCAAGUUUUUAAGU--------ACUACGAGCCAAAUAAAGCUG --.((((((((-----------((..((((.(((((..(((.(((....))).)-))..))))).))))))))))))))............--------......(((........))). ( -26.00) >DroYak_CAF1 20876 117 + 1 --AGAUUGAUCGAUCUCCAUUCGCAUUUCAACACCUCGUAACUAAGCCAUUACUUUGAAAGGUGAUGAAACAAUUAAUCAAGUGCUUCAGUAUUGUACUACUACGAUCCAAAUAA-CCCG --.....(((((.............(((((.(((((..(((.(((....)))..)))..))))).)))))..........(((((.........)))))....))))).......-.... ( -20.00) >consensus __AGAUUGAUCGAUCUCCAUUCGUAUUUCAACACCUCGUAACUAAGUUAUUACUUUGAAAGGUGAUGAAACUAUUAAUCAACUUUUU_AGUAGUCUACUCGUACGAGCCAAAUAAACCUG ...((((((((((.......)))..(((((.(((((..(((...(((....))))))..))))).)))))..)))))))......................................... (-13.62 = -14.36 + 0.74)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:40 2006