| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,391,342 – 12,391,440 |
| Length | 98 |
| Max. P | 0.911343 |

| Location | 12,391,342 – 12,391,440 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 67.38 |
| Mean single sequence MFE | -28.50 |
| Consensus MFE | -11.29 |
| Energy contribution | -11.73 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.40 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911343 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12391342 98 + 27905053 UCAAC--CACCACCUUGUGG---AUUAAUCCA-A----AC-UGAUG--UAAUUUCCU-UC--------GCAGUUAUAUGGCCCCCCGGGACCUCCAGGACCCAUGGGUCCGCCAGGACAC ....(--(((......))))---.....(((.-(----((-((.((--.........-.)--------))))))...((((((....)).......(((((....))))))))))))... ( -29.10) >DroGri_CAF1 41436 105 + 1 UAUCC--UUCU-CCCUCACG-----CAGUACA-A----GU-AGAUGUGUUAUUUCUA-UUAACAAACAAAAGCUUUAUGGACCACCCGGCCCACCUGGUCCGAUGGGCCCACCUGGACAC ..(((--....-(((((...-----..(((.(-(----((-.....(((((......-.))))).......)))))))((((((...((....)))))))))).))).......)))... ( -20.90) >DroEre_CAF1 60178 100 + 1 UCACCAGCACCGCCCUGUGG---ACUAAUCCA-A----AC-CCAUG--UAAUUUCCU-UC--------GCAGUUAUAUGGCCCCCCGGGACCUCCUGGACCAAUGGGUCCGCCGGGACAC ...........((((.((((---(((......-.----..-(((((--(((((....-..--------..))))))))))....(((((....))))).......))))))).))).).. ( -34.00) >DroWil_CAF1 22384 98 + 1 UAGAC--UACUAAGCAGUGG---CUUAAUUUCUA----AU-U---G--UUAUAUUCUUUC-------AACAGUUAUACGGACCACCUGGUCCUCCUGGUCCCAUGGGUCCUCCAGGACAU .....--...(((((....)---)))).....((----((-(---(--((..........-------))))))))...(((((....)))))((((((.((....))....))))))... ( -28.20) >DroYak_CAF1 57465 103 + 1 UCACC--CACCACCCUGUGG---AUUAACCCA-AUGUAACACAAUG--UAAUUUCCU-UC--------GCAGUUAUAUGGCCCCCCGGGACCUCCAGGACCCAUGGGUCCGCCAGGACAC ...((--(.((((...))))---......(((-.((((((....((--..(......-).--------.)))))))))))......))).(((...(((((....)))))...))).... ( -28.40) >DroAna_CAF1 42056 101 + 1 UGACC--CACCACCGUGCGGCCGUUUAACCCUCA----AC-UGACG--UUAAUUUCU--U--------GCAGUUAUACGGCCCCCCAGGACCACCGGGACCCAUGGGCCCGCCCGGACAC .....--.....(((.((((((((.((((...((----(.-.((..--....))..)--)--------)..)))).)))))).....((....))(((.((....))))))).))).... ( -30.40) >consensus UCACC__CACCACCCUGUGG___AUUAAUCCA_A____AC_UGAUG__UAAUUUCCU_UC________GCAGUUAUAUGGCCCCCCGGGACCUCCUGGACCCAUGGGUCCGCCAGGACAC .......................................................................(((...(((.(((...((.((....)).))...))).)))....))).. (-11.29 = -11.73 + 0.45)

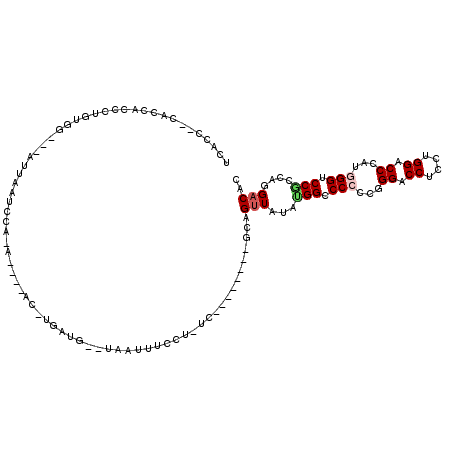

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:21 2006