| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,763,258 – 1,763,362 |
| Length | 104 |
| Max. P | 0.964268 |

| Location | 1,763,258 – 1,763,362 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 74.56 |
| Mean single sequence MFE | -28.00 |
| Consensus MFE | -10.59 |
| Energy contribution | -10.45 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.38 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.964268 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
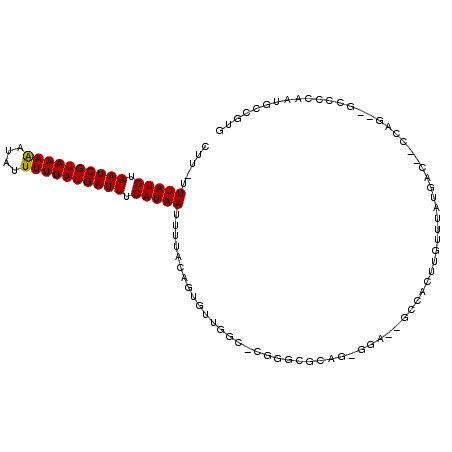
>3R_DroMel_CAF1 1763258 104 + 27905053 CUU-UUUAUGUAAUCGUAAAAAUAUUUUUAUGAUUUCAUAAUUUUACAGUGUUGGU-GGGGCAAAGAGGAAUGCUCCUUGUUUAUAGC--CUCA--GCCACAUUGGCGUG ...-.(((((.((((((((((....)))))))))).))))).....((((((.(((-(((((...(((((....))))).......))--))))--.))))))))..... ( -33.00) >DroPse_CAF1 62517 91 + 1 CUU-UUUAUGUAAUCGUAAAAAUAUUUUUAUGAUUUCAUAAUUUUACAGUGUUGGC-CGGGGGCCGGGGA--GCCACUUGUUUAUGAC--CCAG--GCC----------- ...-.(((((.((((((((((....)))))))))).)))))............(((-(..(((.((..((--((.....)))).)).)--)).)--)))----------- ( -25.40) >DroGri_CAF1 73467 102 + 1 CUU-UUUAUGUAAUCGUAAAAAUAUUUUUAUGAUUUCAUAAUUUUACAGUUUUUGGUUG--CAGA---GA--GUCAGUUGUUUAUGACAUCCGGCAUCCCCCAUAACGUG ...-.(((((.((((((((((....)))))))))).))))).......(((..(((.((--(.(.---((--((((........)))).))).)))....))).)))... ( -22.40) >DroMoj_CAF1 77611 100 + 1 GUUUUUUAUGUAAUCGUAAAGAUAUUUUUAUGAUUUCAUAAUUUUACAGUUUUGGUUUG--CGUA---GA--GGCAGAAGUUUAUGGC--CCUGCAUACCC-AUGGCUGG .....(((((.((((((((((....)))))))))).))))).....((((..(((....--.(((---(.--(((...........))--)))))....))-)..)))). ( -27.10) >DroAna_CAF1 67385 104 + 1 CUU-UUUAUGUAAUCGUAAAAAUAUUUUUAUGAUUUCAUAAUUUUACAGUGUUGGC-GGGGCCGAGAAGAGUGUCGGUUGUUUAUGGC--CAUA--GCCACAUUGCCCUC ...-.(((((.((((((((((....)))))))))).))))).....((((((.(((-..(((((.(((.((......)).))).))))--)...--)))))))))..... ( -32.60) >DroPer_CAF1 69001 102 + 1 CUU-UUUAUGUAAUCGUAAAAAUAUUUUUAUGAUUUCAUAAUUUUACAGUGUUCGC-CGGGGGCCGGGGA--GCCACUUGUUUAUGAC--CCAG--GGCCCAGUACCUUG ...-.(((((.((((((((((....)))))))))).)))))....(((((((((.(-(((...)))).))--).))).))).......--.(((--((.......))))) ( -27.50) >consensus CUU_UUUAUGUAAUCGUAAAAAUAUUUUUAUGAUUUCAUAAUUUUACAGUGUUGGC_CGGGCGCAG_GGA__GCCACUUGUUUAUGAC__CCAG__GCCCCAAUGCCGUG .....(((((.((((((((((....)))))))))).)))))..................................................................... (-10.59 = -10.45 + -0.14)

| Location | 1,763,258 – 1,763,362 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 74.56 |
| Mean single sequence MFE | -21.77 |
| Consensus MFE | -6.53 |
| Energy contribution | -6.70 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.30 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834356 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
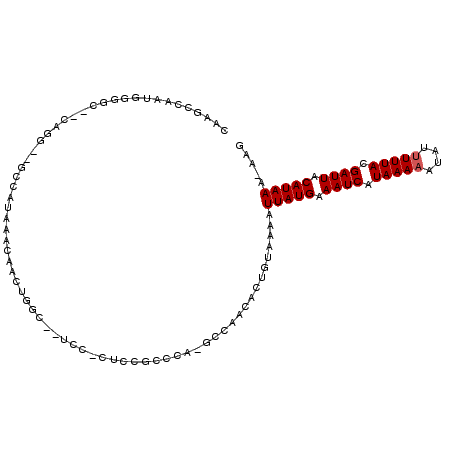
>3R_DroMel_CAF1 1763258 104 - 27905053 CACGCCAAUGUGGC--UGAG--GCUAUAAACAAGGAGCAUUCCUCUUUGCCCC-ACCAACACUGUAAAAUUAUGAAAUCAUAAAAAUAUUUUUACGAUUACAUAAA-AAG .....((.(((((.--((.(--((.......((((((.....))))))))).)-))).))).)).....(((((.((((.(((((....))))).)))).))))).-... ( -19.11) >DroPse_CAF1 62517 91 - 1 -----------GGC--CUGG--GUCAUAAACAAGUGGC--UCCCCGGCCCCCG-GCCAACACUGUAAAAUUAUGAAAUCAUAAAAAUAUUUUUACGAUUACAUAAA-AAG -----------(((--(.((--(((((......)))))--))...((...)))-)))............(((((.((((.(((((....))))).)))).))))).-... ( -22.10) >DroGri_CAF1 73467 102 - 1 CACGUUAUGGGGGAUGCCGGAUGUCAUAAACAACUGAC--UC---UCUG--CAACCAAAAACUGUAAAAUUAUGAAAUCAUAAAAAUAUUUUUACGAUUACAUAAA-AAG ....((((((.((.(((.((..((((........))))--..---)).)--)).)).....))))))..(((((.((((.(((((....))))).)))).))))).-... ( -20.20) >DroMoj_CAF1 77611 100 - 1 CCAGCCAU-GGGUAUGCAGG--GCCAUAAACUUCUGCC--UC---UACG--CAAACCAAAACUGUAAAAUUAUGAAAUCAUAAAAAUAUCUUUACGAUUACAUAAAAAAC .(((....-.(((.((((((--((...........)))--.)---)..)--)).)))....))).....(((((.((((.((((......)))).)))).)))))..... ( -18.20) >DroAna_CAF1 67385 104 - 1 GAGGGCAAUGUGGC--UAUG--GCCAUAAACAACCGACACUCUUCUCGGCCCC-GCCAACACUGUAAAAUUAUGAAAUCAUAAAAAUAUUUUUACGAUUACAUAAA-AAG ...(((..((((((--....--)))))).....((((........))))....-)))............(((((.((((.(((((....))))).)))).))))).-... ( -24.30) >DroPer_CAF1 69001 102 - 1 CAAGGUACUGGGCC--CUGG--GUCAUAAACAAGUGGC--UCCCCGGCCCCCG-GCGAACACUGUAAAAUUAUGAAAUCAUAAAAAUAUUUUUACGAUUACAUAAA-AAG ....((...(((((--..((--(((((......)))))--.))..)))))...-)).............(((((.((((.(((((....))))).)))).))))).-... ( -26.70) >consensus CAAGCCAAUGGGGC__CAGG__GCCAUAAACAACUGGC__UCC_CUCCGCCCA_GCCAACACUGUAAAAUUAUGAAAUCAUAAAAAUAUUUUUACGAUUACAUAAA_AAG .....................................................................(((((.((((.(((((....))))).)))).)))))..... ( -6.53 = -6.70 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:02 2006