
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,385,848 – 12,385,941 |
| Length | 93 |
| Max. P | 0.758862 |

| Location | 12,385,848 – 12,385,941 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 84.78 |
| Mean single sequence MFE | -24.78 |
| Consensus MFE | -17.16 |
| Energy contribution | -18.04 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.510515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
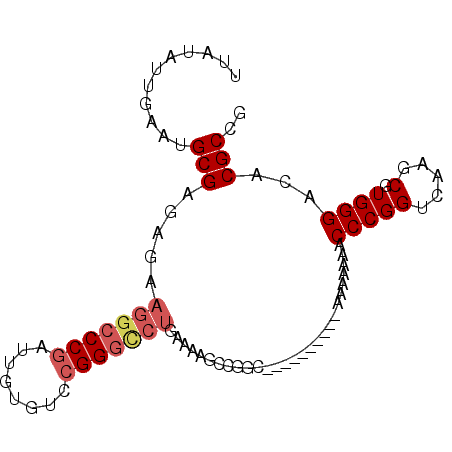
>3R_DroMel_CAF1 12385848 93 + 27905053 UUAUAUUGAAUGCGAGAGAAGGCCCGAUUGUGUCCGGGCCUGAAAAGCCGGCAAAAAAAAAAAAAAAAACCCGGUCAAGCGUGGGACACGCCG .....((((..........(((((((........)))))))......((((...................))))))))(((((...))))).. ( -24.81) >DroSec_CAF1 54286 82 + 1 UUAUAUUGAAUGCGAGAGAAGGCCCGAUUGUGUACGGGUCUGAAAAGCCGG-----------AAAAAAACCCGGUCAAGCGUGGGACACGCCG .....((((..........(((((((........)))))))......((((-----------........))))))))(((((...))))).. ( -24.10) >DroSim_CAF1 53634 82 + 1 UUAUAUUGAAUGCGAGAGAAGGCCCGAUUGUGUACGGGCCUGAAAAGCCGG-----------AAAAAAACCCGGUCAAGCGUGGGACACGCCG .....((((..........(((((((........)))))))......((((-----------........))))))))(((((...))))).. ( -26.80) >DroEre_CAF1 54966 71 + 1 UUAUAUUGAAUGCGAGAGAAAGCCCGCUUGUGACCGG-------------GCC---------AAAAAAACCCGGUCAAGCGUGGGACACGCCG ...........(((.(......(((((...(((((((-------------(..---------.......))))))))...))))).).))).. ( -25.20) >DroYak_CAF1 51729 84 + 1 UUAUAUUGAAUGCGAGAGAAGGCCCGUUUGUGUCAGGGCCUGAAAACCCGGCA---------AAAAAAACCCGGUCAAGCGUGGGACACGCCG .....((((..........((((((..........))))))......((((..---------........))))))))(((((...))))).. ( -23.00) >consensus UUAUAUUGAAUGCGAGAGAAGGCCCGAUUGUGUCCGGGCCUGAAAAGCCGGC__________AAAAAAACCCGGUCAAGCGUGGGACACGCCG ...........(((.....(((((((........)))))))............................(((((.....).))))...))).. (-17.16 = -18.04 + 0.88)


| Location | 12,385,848 – 12,385,941 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 84.78 |
| Mean single sequence MFE | -25.72 |
| Consensus MFE | -16.03 |
| Energy contribution | -17.83 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.758862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12385848 93 - 27905053 CGGCGUGUCCCACGCUUGACCGGGUUUUUUUUUUUUUUUUUGCCGGCUUUUCAGGCCCGGACACAAUCGGGCCUUCUCUCGCAUUCAAUAUAA ..((((((((...((((((((((...................))))....))))))..))))))....(((......)))))........... ( -24.81) >DroSec_CAF1 54286 82 - 1 CGGCGUGUCCCACGCUUGACCGGGUUUUUUU-----------CCGGCUUUUCAGACCCGUACACAAUCGGGCCUUCUCUCGCAUUCAAUAUAA .((((((...))))))((((((((......)-----------))))......((.((((........)))).))..........)))...... ( -20.60) >DroSim_CAF1 53634 82 - 1 CGGCGUGUCCCACGCUUGACCGGGUUUUUUU-----------CCGGCUUUUCAGGCCCGUACACAAUCGGGCCUUCUCUCGCAUUCAAUAUAA .((((((...))))))((((((((......)-----------))))......(((((((........)))))))..........)))...... ( -27.70) >DroEre_CAF1 54966 71 - 1 CGGCGUGUCCCACGCUUGACCGGGUUUUUUU---------GGC-------------CCGGUCACAAGCGGGCUUUCUCUCGCAUUCAAUAUAA .((((((...))))))((((((((((.....---------)))-------------)))))))...(((((......)))))........... ( -30.20) >DroYak_CAF1 51729 84 - 1 CGGCGUGUCCCACGCUUGACCGGGUUUUUUU---------UGCCGGGUUUUCAGGCCCUGACACAAACGGGCCUUCUCUCGCAUUCAAUAUAA .((((((...)))))).((((.(((......---------.))).))))...((((((..........))))))................... ( -25.30) >consensus CGGCGUGUCCCACGCUUGACCGGGUUUUUUU__________GCCGGCUUUUCAGGCCCGGACACAAUCGGGCCUUCUCUCGCAUUCAAUAUAA .((((((...))))))(((((((...................))))......(((((((........)))))))..........)))...... (-16.03 = -17.83 + 1.80)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:15 2006