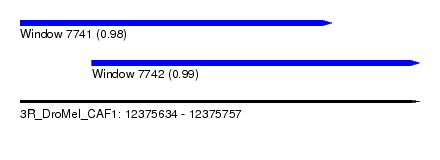
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,375,634 – 12,375,757 |
| Length | 123 |
| Max. P | 0.994218 |

| Location | 12,375,634 – 12,375,730 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 88.77 |
| Mean single sequence MFE | -29.32 |
| Consensus MFE | -25.30 |
| Energy contribution | -25.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984544 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
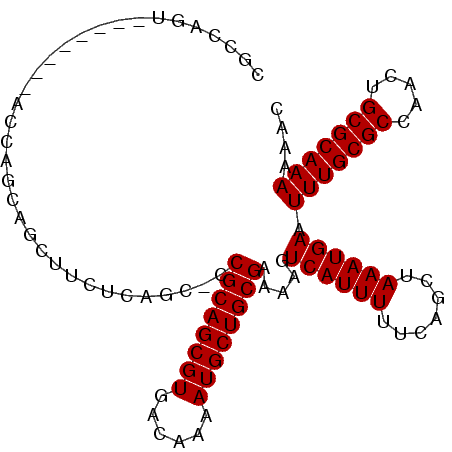
>3R_DroMel_CAF1 12375634 96 + 27905053 CGCCAUU-------CGCCAGCAGCUUCUCAGC-CCGCAGCGUGACAAAAUGCUGCGAAAAGUCAUUUUUCAGCUAAAUGAAUUUGCGCCAACUGCGCAAAAAAC .....((-------((..(((.(((....)))-.((((((((......))))))))...............)))...))))(((((((.....))))))).... ( -27.70) >DroSec_CAF1 44030 95 + 1 CGCCAGC--------AGCAGCAGCUUCUCAGC-CCGCAGCGUGACAAAAUGCUGCGAAAAGUCAUUUUUCAGCUAAAUGAAUUUGCGCCAACUGCGCAAAAAAC .((.((.--------(((....))).))..))-.((((((((......)))))))).....((((((.......)))))).(((((((.....))))))).... ( -29.20) >DroSim_CAF1 43383 95 + 1 CGCCAGC--------AGCAGCAGCUUCUCAGC-CCGCAGCGUGACAAAAUGCUGCGAAAAGUCAUUUUUCAGCUAAAUGAAUUUGCGCCAAUUGCGCAAAAAAC .((.((.--------(((....))).))..))-.((((((((......)))))))).....((((((.......)))))).(((((((.....))))))).... ( -29.20) >DroEre_CAF1 44397 103 + 1 CGCCACUAGCUAUACGCCAGCAGAUUCUCAGC-CCGCAGCGUGACAAAAUGCUGCGAAAAGUCAUUUUUCAGCUAAAUGAAUUUGCGCCAACUGCGCAAAAAAG .((.....)).....((..((((((((..(((-..(((((((......)))))))((((((...)))))).)))....))))))))((.....))))....... ( -28.70) >DroYak_CAF1 40949 103 + 1 CGCCAUUCGCUAUUCGCCAGCAGCUUCUCAGC-CCGCAGCGUGACAAAAUGCUGCGAAAAGUCAUUUUUCAGCUAAAUGAAUUUGCGCCAACUGCGCAAAAAAC ...((((.(((.......)))((((.......-..(((((((......)))))))((((((...)))))))))).))))..(((((((.....))))))).... ( -28.30) >DroAna_CAF1 25759 93 + 1 CGCUGGG--------A---GCUUCUUCUCUCCGCCGCAGCGUGACAAAAUGCUGCGAAAAGUCAUUUUUCAGCUAAAUGAAUUUGCGCCAACUGCGCAAAAAAC .((((((--------(---(........))))..((((((((......)))))))).............))))........(((((((.....))))))).... ( -32.80) >consensus CGCCAGU________ACCAGCAGCUUCUCAGC_CCGCAGCGUGACAAAAUGCUGCGAAAAGUCAUUUUUCAGCUAAAUGAAUUUGCGCCAACUGCGCAAAAAAC ..................................((((((((......)))))))).....((((((.......)))))).(((((((.....))))))).... (-25.30 = -25.30 + 0.00)

| Location | 12,375,656 – 12,375,757 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 87.39 |
| Mean single sequence MFE | -29.75 |
| Consensus MFE | -25.30 |
| Energy contribution | -25.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.06 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.994218 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12375656 101 + 27905053 AG--C-CCGCAGCGUGACAAAAUGCUGCGAAAAGUCAUUUUUCAGCUAAAUGAAUUUGCGCCAACUGCGCAAAAAACAACGC--CCAAAAAAGUAU--CUUAACAGCA .(--(-.((((((((......)))))))).....((((((.......)))))).(((((((.....))))))).........--............--.......)). ( -26.60) >DroPse_CAF1 41170 99 + 1 CGUCC-CCGCAGCGUGACAAAAUGCUGCGAAAAGUCAUUUUUCAGCUAAAUGAAUUUGCGCCAACUGCGCAAAAAACAACGA--CCAAGAAGAUGG--C----CAGAA ((((.-.((((((((......))))))))....(((....((((......))))(((((((.....))))))).......))--)......)))).--.----..... ( -29.80) >DroEre_CAF1 44426 103 + 1 AG--C-CCGCAGCGUGACAAAAUGCUGCGAAAAGUCAUUUUUCAGCUAAAUGAAUUUGCGCCAACUGCGCAAAAAAGAACGC--CCAAAAAAGUGGCGCUUAACUGCA ..--.-.((((((((......))))))))....(((((((((((......))).(((((((.....))))))).........--.....))))))))((......)). ( -32.30) >DroYak_CAF1 40978 101 + 1 AG--C-CCGCAGCGUGACAAAAUGCUGCGAAAAGUCAUUUUUCAGCUAAAUGAAUUUGCGCCAACUGCGCAAAAAACAACGC--CCAAAAAAGUGG--CUAAACUGCA .(--(-.((((((((......))))))))...((((((((((((......))).(((((((.....))))))).........--.....)))))))--)).....)). ( -32.50) >DroAna_CAF1 25777 101 + 1 UC--CGCCGCAGCGUGACAAAAUGCUGCGAAAAGUCAUUUUUCAGCUAAAUGAAUUUGCGCCAACUGCGCAAAAAACAACCCGACGAAAAAAAAAU--UAUAA---AA ..--...((((((((......))))))))....(((....((((......))))(((((((.....))))))).........)))...........--.....---.. ( -27.80) >DroPer_CAF1 38253 99 + 1 CGUCC-CCGCAGCGUGACAAAAUGCUGCGAAAAGUCAUUUUUCAGCUAAAUGAAUUUGCGCCAACUGCGCAAAAAACAACGA--CCAAGAAAAUGG--C----CAGAA .....-.((((((((......))))))))....(((((((((((......))).(((((((.....))))))).........--.....)))))))--)----..... ( -29.50) >consensus AG__C_CCGCAGCGUGACAAAAUGCUGCGAAAAGUCAUUUUUCAGCUAAAUGAAUUUGCGCCAACUGCGCAAAAAACAACGC__CCAAAAAAAUGG__CUUAACAGAA .......((((((((......)))))))).....((((((.......)))))).(((((((.....)))))))................................... (-25.30 = -25.30 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:10 2006