
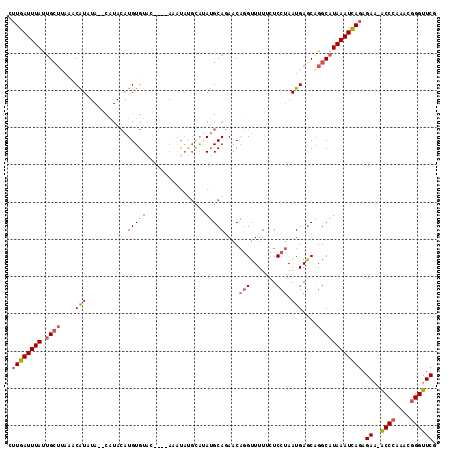
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,371,487 – 12,371,617 |
| Length | 130 |
| Max. P | 0.979146 |

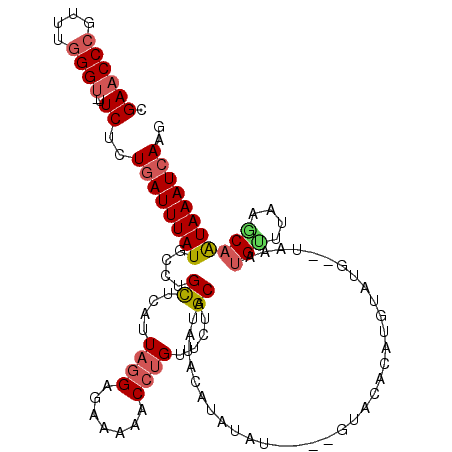
| Location | 12,371,487 – 12,371,596 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 75.91 |
| Mean single sequence MFE | -28.20 |
| Consensus MFE | -10.67 |
| Energy contribution | -11.79 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.38 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979146 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12371487 109 + 27905053 CUUGAUUUAUUGCUUAAACAUUUA--CAUACAUGUGUACACACAAA---CUGAAUGCAGAACAGGUUUUGCUCCUAAUGAGCGGGCAUAAAUCAGAGAA-ACCCAAACGGGUUCG .(((((((((.((((((((.....--......((((....))))..---(((....))).....)))).((((.....))))))))))))))))).(((-.(((....)))))). ( -27.60) >DroSim_CAF1 39189 112 + 1 CUUGAUUUAUUGCUUACACAUUUA--CAUACAUGUGUACACAUACGUAUGUAUAUGCAGAACAGGUUUUUCUCCUAAUGAACAGGCAUAAAUCAGAGAA-ACCCAAACGGGUUCG .(((((((((.((((...(((.((--(((((.((((....)))).))))))).)))......(((.......))).......))))))))))))).(((-.(((....)))))). ( -31.20) >DroEre_CAF1 40182 108 + 1 GUUGAUUUACUGAUGUACCAUAUAUAUAUACAUGUGU------AUAUACGAGUAUGCAGAACAGGUUUUUCUCCUAAUGAGCAGGCAUAAAUCAGAGAA-ACCCAAACGGGUUCG .(((((((((((.((((((.(((((((((....))))------))))).).)))))))).....((((..(((.....))).)))).)))))))).(((-.(((....)))))). ( -25.90) >DroYak_CAF1 36560 109 + 1 CUUGAUUUACUGCUUGAGCAUAUA--CAUACAUGUGUAC---UAUAUAUGCAUAUGCAGAACAGGUUUUACUCCUAAUGAGCAGGCAUAAAUCAGAGAA-GCCCAAACGGGUUCG .((((((((.((((((..(((...--....((((((((.---......))))))))......(((.......))).)))..)))))))))))))).(((-.(((....)))))). ( -31.60) >DroAna_CAF1 21680 96 + 1 ACUGAUUUAUGGCAGA--CGU-----------UGUGUA------AAUAUGCAGAGGCAUAACCUGUUUUCGUCCUAAUGAGCAGGCAUAAAUUACAGAGGACCGAAACGGGUUCU .((((((((((.....--...-----------......------..(((((....))))).(((((((..........))))))))))))))..)))((((((......)))))) ( -24.70) >consensus CUUGAUUUAUUGCUUAAACAUAUA__CAUACAUGUGUAC____AAAUAUGCAUAUGCAGAACAGGUUUUUCUCCUAAUGAGCAGGCAUAAAUCAGAGAA_ACCCAAACGGGUUCG .((((((((.((((....(((...........(((((................)))))....(((.......))).)))....)))))))))))).((..((((....)))))). (-10.67 = -11.79 + 1.12)


| Location | 12,371,487 – 12,371,596 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 75.91 |
| Mean single sequence MFE | -27.50 |
| Consensus MFE | -10.22 |
| Energy contribution | -10.90 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.37 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.953809 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12371487 109 - 27905053 CGAACCCGUUUGGGU-UUCUCUGAUUUAUGCCCGCUCAUUAGGAGCAAAACCUGUUCUGCAUUCAG---UUUGUGUGUACACAUGUAUG--UAAAUGUUUAAGCAAUAAAUCAAG .((((((....))))-))...((((((((((.....((((.((((((.....))))))((((.((.---..((((....)))))).)))--).)))).....)).)))))))).. ( -27.20) >DroSim_CAF1 39189 112 - 1 CGAACCCGUUUGGGU-UUCUCUGAUUUAUGCCUGUUCAUUAGGAGAAAAACCUGUUCUGCAUAUACAUACGUAUGUGUACACAUGUAUG--UAAAUGUGUAAGCAAUAAAUCAAG .((((((....))))-))...((((((((.((((.....))))..............(((.((((((((((((((((....))))))))--))..)))))).))))))))))).. ( -33.20) >DroEre_CAF1 40182 108 - 1 CGAACCCGUUUGGGU-UUCUCUGAUUUAUGCCUGCUCAUUAGGAGAAAAACCUGUUCUGCAUACUCGUAUAU------ACACAUGUAUAUAUAUAUGGUACAUCAGUAAAUCAAC .((((((....))))-))..(((((...(((((((....((((.......))))....))).....((((((------((....))))))))....)))).)))))......... ( -26.20) >DroYak_CAF1 36560 109 - 1 CGAACCCGUUUGGGC-UUCUCUGAUUUAUGCCUGCUCAUUAGGAGUAAAACCUGUUCUGCAUAUGCAUAUAUA---GUACACAUGUAUG--UAUAUGCUCAAGCAGUAAAUCAAG .((((((....))).-)))..(((((((....(((((.....)))))....(((((..((((((((((((((.---(....))))))))--)))))))...)))))))))))).. ( -35.30) >DroAna_CAF1 21680 96 - 1 AGAACCCGUUUCGGUCCUCUGUAAUUUAUGCCUGCUCAUUAGGACGAAAACAGGUUAUGCCUCUGCAUAUU------UACACA-----------ACG--UCUGCCAUAAAUCAGU ((((((.((((..(((((..(((.........))).....)))))..)))).)))(((((....)))))..------......-----------...--)))............. ( -15.60) >consensus CGAACCCGUUUGGGU_UUCUCUGAUUUAUGCCUGCUCAUUAGGAGAAAAACCUGUUCUGCAUAUACAUAUAU____GUACACAUGUAUG__UAAAUGUUUAAGCAAUAAAUCAAG .((((((....))))..))..((((((((....((....((((.......))))....))...................................(((....))))))))))).. (-10.22 = -10.90 + 0.68)

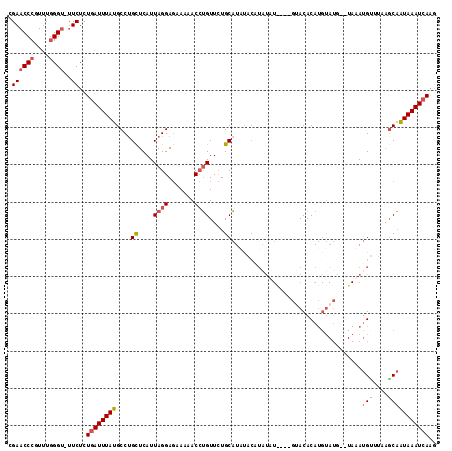
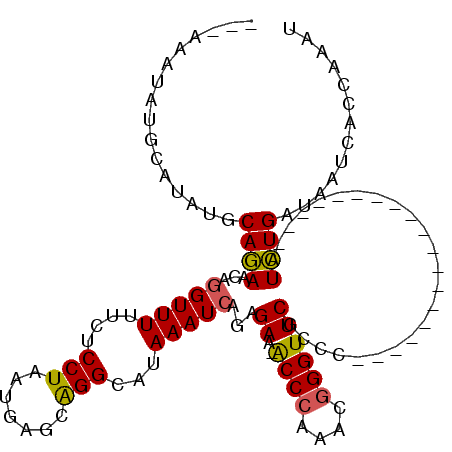
| Location | 12,371,525 – 12,371,617 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 77.61 |
| Mean single sequence MFE | -23.46 |
| Consensus MFE | -10.80 |
| Energy contribution | -10.56 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12371525 92 + 27905053 CACAAA---CUGAAUGCAGAACAGGUUUUGCUCCUAAUGAGCGGGCAUAAAUCAGAGAA-ACCCAAACGGGUUCGCCC-------------------AUCUGAUAAUCACCAAAU ......---(((....)))..(((((...((((.....))))((((.....((...))(-((((....))))).))))-------------------)))))............. ( -22.50) >DroSim_CAF1 39227 95 + 1 CAUACGUAUGUAUAUGCAGAACAGGUUUUUCUCCUAAUGAACAGGCAUAAAUCAGAGAA-ACCCAAACGGGUUCGCCC-------------------AUCUGAUAAUCACCAAAU .((((....))))...((((...(((((((((....(((......))).....))))))-))).....(((....)))-------------------.))))............. ( -19.20) >DroEre_CAF1 40219 92 + 1 ---AUAUACGAGUAUGCAGAACAGGUUUUUCUCCUAAUGAGCAGGCAUAAAUCAGAGAA-ACCCAAACGGGUUCGCCC-------------------AUCUGAUAAUCACCAAAU ---......((.(((.((((...(((((((((....(((......))).....))))))-))).....(((....)))-------------------.))))))).))....... ( -21.00) >DroYak_CAF1 36597 93 + 1 --UAUAUAUGCAUAUGCAGAACAGGUUUUACUCCUAAUGAGCAGGCAUAAAUCAGAGAA-GCCCAAACGGGUUCGCCC-------------------AUCUGAUAAUCACCAAAU --....(((((...(((.....(((.......))).....))).))))).((((((..(-((((....))))).....-------------------.))))))........... ( -22.20) >DroAna_CAF1 21705 111 + 1 ----AAUAUGCAGAGGCAUAACCUGUUUUCGUCCUAAUGAGCAGGCAUAAAUUACAGAGGACCGAAACGGGUUCUCCCUCUCCCUCUCCGUCUCUGCCUGUGAUAAGCACCAAAU ----.(((.((((((((....(((((((..........)))))))...........(((((((......))))))).............)))))))).))).............. ( -32.40) >consensus ___AAAUAUGCAUAUGCAGAACAGGUUUUUCUCCUAAUGAGCAGGCAUAAAUCAGAGAA_ACCCAAACGGGUUCGCCC___________________AUCUGAUAAUCACCAAAU ................((((...(((((....(((.......)))...)))))...((..((((....))))))........................))))............. (-10.80 = -10.56 + -0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:07 2006