
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
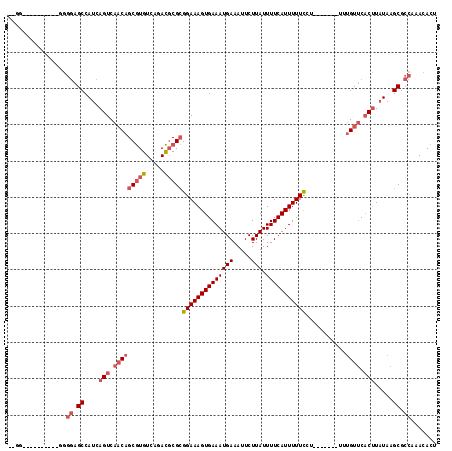
| Location | 12,368,925 – 12,369,030 |
| Length | 105 |
| Max. P | 0.834318 |

| Location | 12,368,925 – 12,369,030 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 82.92 |
| Mean single sequence MFE | -27.25 |
| Consensus MFE | -17.35 |
| Energy contribution | -18.63 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12368925 105 + 27905053 --GA----------GGGGAGCCAUCAGUCAACAGCGUGUCAGACGCGCGGAAAGUGAAAUGAAAUUCUUAUUUUCAUUUUUUUU-UUUUUUUUUGUUCACUUAUAAGCGCCAAACACU --..----------..((.((....(((.(((((((((.....)))))((((((.(((((((((.......))))))))).)))-))).....)))).))).....)).))....... ( -24.60) >DroSec_CAF1 37309 99 + 1 ---------------GGGAGCCAUCAGUCAACAGCGUGUCAGACGCGCGGAAAGUGAAAUGAAAUUCUUAUUUUCAUUUUUCCUUUU----UUUGUUCACUUAUAAGCGCCAAACACU ---------------.((.((....(((.(((((((((.....)))))((((((.(((((((((.......)))))))))..)))))----).)))).))).....)).))....... ( -25.30) >DroSim_CAF1 36612 103 + 1 ---------------GGGAGCCAUCAGUCAACAGCGUGUCAGACGCGCGGAAAGUGAAAUGAAAUUCUUAUUUUCAUUUUUCCUUUUUUUUUUUGUUCACUUAUAAGCGCCAAACACU ---------------.((.((....(((.(((((((((.....)))))((((((.(((((((((.......)))))))))..)))))).....)))).))).....)).))....... ( -25.30) >DroEre_CAF1 37618 110 + 1 GUGGGGGGAGCCAGGGGGAGCCAUCAGUCAACAGCGUGUCAGACGCGCGGAAAGUGAAAUGAAAUUCUUAUUUUCAUUUUUCCA--------CUGUUCACUUAUAAGCGCCAAACACU (((...((.((...((....))...(((.(((((((((.....)))))(((....(((((((((.......)))))))))))).--------.)))).))).....)).))...))). ( -31.40) >DroYak_CAF1 33889 97 + 1 GUGG----------GGGGAGCCAUCAGUCAACAGCGUGUCAGACGCGCGGAAAGUGAAAUGAAAUUCUUAUUUUCAUUUUUU-----------UGUUCACUUAUAAGCGCCAAACACU (((.----------..((.((....(((.(((((((((.....)))))((((((((((((((.....))).)))))))))))-----------)))).))).....)).))...))). ( -26.80) >DroAna_CAF1 19139 101 + 1 --GG----------CAACAGCCAUCAGUCAACAGCAUGUCAGAUCCGAGGAAAGUGAAAUGAAAUUCUUAUUUUCAUUUUUU-----UCCUCUGGCGCCCAUCUAAGCGCCAAACACU --((----------(....)))........................(((((((..(((((((((.......)))))))))))-----)))))((((((........))))))...... ( -30.10) >consensus __GG__________GGGGAGCCAUCAGUCAACAGCGUGUCAGACGCGCGGAAAGUGAAAUGAAAUUCUUAUUUUCAUUUUUCCU_______UUUGUUCACUUAUAAGCGCCAAACACU ................((.((....(((.(((((((((.....)))))((((((((((((((.....))).)))))))))))...........)))).))).....)).))....... (-17.35 = -18.63 + 1.28)


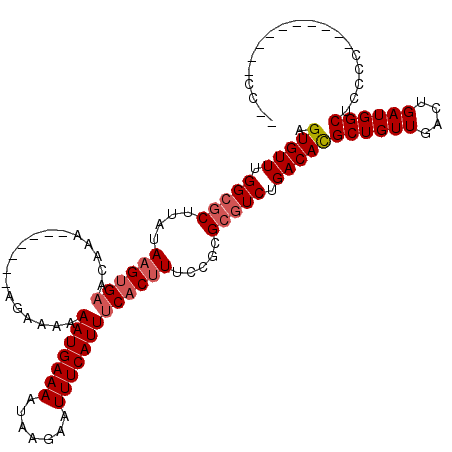
| Location | 12,368,925 – 12,369,030 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 82.92 |
| Mean single sequence MFE | -28.67 |
| Consensus MFE | -21.63 |
| Energy contribution | -22.33 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834318 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12368925 105 - 27905053 AGUGUUUGGCGCUUAUAAGUGAACAAAAAAAAA-AAAAAAAAUGAAAAUAAGAAUUUCAUUUCACUUUCCGCGCGUCUGACACGCUGUUGACUGAUGGCUCCCC----------UC-- .(((((.(((((....(((((((..........-.......((((((.......))))))))))))).....))))).)))))((((((....)))))).....----------..-- ( -25.92) >DroSec_CAF1 37309 99 - 1 AGUGUUUGGCGCUUAUAAGUGAACAAA----AAAAGGAAAAAUGAAAAUAAGAAUUUCAUUUCACUUUCCGCGCGUCUGACACGCUGUUGACUGAUGGCUCCC--------------- .(((((.(((((......(....)...----....((((((((((((.......)))))))....)))))..))))).)))))((((((....))))))....--------------- ( -27.00) >DroSim_CAF1 36612 103 - 1 AGUGUUUGGCGCUUAUAAGUGAACAAAAAAAAAAAGGAAAAAUGAAAAUAAGAAUUUCAUUUCACUUUCCGCGCGUCUGACACGCUGUUGACUGAUGGCUCCC--------------- .(((((.(((((......(....)...........((((((((((((.......)))))))....)))))..))))).)))))((((((....))))))....--------------- ( -27.00) >DroEre_CAF1 37618 110 - 1 AGUGUUUGGCGCUUAUAAGUGAACAG--------UGGAAAAAUGAAAAUAAGAAUUUCAUUUCACUUUCCGCGCGUCUGACACGCUGUUGACUGAUGGCUCCCCCUGGCUCCCCCCAC .(((((.(((((......(....).(--------(((((((((((((.......)))))))....)))))))))))).)))))((((((....))))))......(((......))). ( -31.70) >DroYak_CAF1 33889 97 - 1 AGUGUUUGGCGCUUAUAAGUGAACA-----------AAAAAAUGAAAAUAAGAAUUUCAUUUCACUUUCCGCGCGUCUGACACGCUGUUGACUGAUGGCUCCCC----------CCAC .(((((.(((((....(((((((..-----------.....((((((.......))))))))))))).....))))).)))))((((((....)))))).....----------.... ( -26.81) >DroAna_CAF1 19139 101 - 1 AGUGUUUGGCGCUUAGAUGGGCGCCAGAGGA-----AAAAAAUGAAAAUAAGAAUUUCAUUUCACUUUCCUCGGAUCUGACAUGCUGUUGACUGAUGGCUGUUG----------CC-- ......((((((((....))))))))(((((-----((.((((((((.......))))))))...)))))))((....((((.((((((....)))))))))).----------))-- ( -33.60) >consensus AGUGUUUGGCGCUUAUAAGUGAACAAA_______AGAAAAAAUGAAAAUAAGAAUUUCAUUUCACUUUCCGCGCGUCUGACACGCUGUUGACUGAUGGCUCCCC__________CC__ .(((((.(((((....((((((..................(((((((.......))))))))))))).....))))).)))))((((((....))))))................... (-21.63 = -22.33 + 0.70)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:04 2006