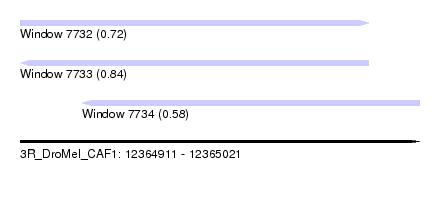
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,364,911 – 12,365,021 |
| Length | 110 |
| Max. P | 0.841463 |

| Location | 12,364,911 – 12,365,007 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.83 |
| Mean single sequence MFE | -28.55 |
| Consensus MFE | -19.09 |
| Energy contribution | -20.32 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.723322 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12364911 96 + 27905053 -----------------------GCUGGCAAAUCAUGCUGUCUAA-AUUUGUAUUUAUUAGACAAUGCGCCACGGGGCGUAUGUGUAAUAUGCCAUGCUAUGCCAUUAGAUGCAUUCCAA -----------------------(((((((.((.(((((((((((-...........)))))))(((((((....))))))))))).)).))))).)).((((........))))..... ( -28.40) >DroPse_CAF1 31970 119 + 1 UCUGGCUACGAGGCGGACGUUGCUCUGGCAAAUCAUGCUGUCUAA-AUUUGUAUUUAUUAGACAAUGCGCCACGGGGCGUAUGUGUAAUAUGCCAUGCCAUGCCAUUAGAUGCAUUCCAG (((((((....)))((.(((.((..(((((.((.(((((((((((-...........)))))))(((((((....))))))))))).)).))))).)).)))))..)))).......... ( -35.00) >DroGri_CAF1 14481 100 + 1 A--GGGC--------AAAGUUCCGCUGGCAAUUCAUGCUGUCCAAAAUUUGUAUUUAUUAGACAGAACGCCACGGGGCGUAUGUGUAUUAUG----------CCAAUAUAAGCAUUCCAA .--((((--------......(((.((((........(((((..((((....))))....)))))...)))))))((((((.......))))----------)).......))...)).. ( -23.90) >DroYak_CAF1 29854 96 + 1 -----------------------GCUGGCAAAUCAUGCUGUCUAA-AUUUGUAUUUAUUAGAAAAUGCGCCACGGGGCGUAUGUGUAAUAUGCAAUGCUAUGCCAUUAGAUGCAUUCCAA -----------------------((.((((.....))))((((((-.........(((((.(..(((((((....))))))).).))))).(((......)))..))))))))....... ( -21.70) >DroAna_CAF1 15718 94 + 1 -------------------------CGGCAAAUCAUGCUGUCUAA-AUUUGUAUUUAUUAGACAAUGCGCCACAGGGCGUAUGUGUAAUAUGCCAUGCUAUGCCAUUAGAUGCAUUCCAA -------------------------.((((.((.(((((((((((-...........)))))))(((((((....))))))))))).)).)))).((..((((........))))..)). ( -27.30) >DroPer_CAF1 29071 119 + 1 UCUGGCUACGAGGCGGACGUUGCUCUGGCAAAUCAUGCUGUCUAA-AUUUGUAUUUAUUAGACAAUGCGCCACGGGGCGUAUGUGUAAUAUGCCAUGCCAUGCCAUUAGAUGCAUUCCAG (((((((....)))((.(((.((..(((((.((.(((((((((((-...........)))))))(((((((....))))))))))).)).))))).)).)))))..)))).......... ( -35.00) >consensus _______________________GCUGGCAAAUCAUGCUGUCUAA_AUUUGUAUUUAUUAGACAAUGCGCCACGGGGCGUAUGUGUAAUAUGCCAUGCUAUGCCAUUAGAUGCAUUCCAA ..........................((((...((((((((((((.(........).)))))))(((((((....))))))).........)).)))...))))................ (-19.09 = -20.32 + 1.22)
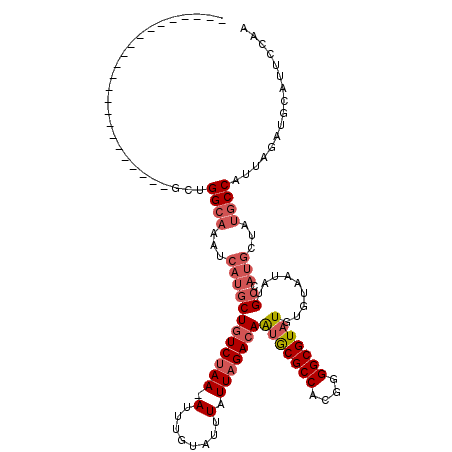
| Location | 12,364,911 – 12,365,007 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.83 |
| Mean single sequence MFE | -26.78 |
| Consensus MFE | -18.45 |
| Energy contribution | -19.37 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841463 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12364911 96 - 27905053 UUGGAAUGCAUCUAAUGGCAUAGCAUGGCAUAUUACACAUACGCCCCGUGGCGCAUUGUCUAAUAAAUACAAAU-UUAGACAGCAUGAUUUGCCAGC----------------------- (((((.....)))))(((((...((((..............((((....))))..((((((((...........-))))))))))))...)))))..----------------------- ( -25.60) >DroPse_CAF1 31970 119 - 1 CUGGAAUGCAUCUAAUGGCAUGGCAUGGCAUAUUACACAUACGCCCCGUGGCGCAUUGUCUAAUAAAUACAAAU-UUAGACAGCAUGAUUUGCCAGAGCAACGUCCGCCUCGUAGCCAGA ((((..(((......(((((...((((..............((((....))))..((((((((...........-))))))))))))...)))))((((.......).)))))).)))). ( -33.00) >DroGri_CAF1 14481 100 - 1 UUGGAAUGCUUAUAUUGG----------CAUAAUACACAUACGCCCCGUGGCGUUCUGUCUAAUAAAUACAAAUUUUGGACAGCAUGAAUUGCCAGCGGAACUUU--------GCCC--U ..((.(((((......))----------)))..............((((((((..((((((((............)))))))).......))))).)))......--------..))--. ( -23.90) >DroYak_CAF1 29854 96 - 1 UUGGAAUGCAUCUAAUGGCAUAGCAUUGCAUAUUACACAUACGCCCCGUGGCGCAUUUUCUAAUAAAUACAAAU-UUAGACAGCAUGAUUUGCCAGC----------------------- (((((.....)))))(((((...(((.((............((((....)))).....(((((...........-)))))..)))))...)))))..----------------------- ( -20.60) >DroAna_CAF1 15718 94 - 1 UUGGAAUGCAUCUAAUGGCAUAGCAUGGCAUAUUACACAUACGCCCUGUGGCGCAUUGUCUAAUAAAUACAAAU-UUAGACAGCAUGAUUUGCCG------------------------- (((((.....))))).((((...((((..............((((....))))..((((((((...........-))))))))))))...)))).------------------------- ( -24.60) >DroPer_CAF1 29071 119 - 1 CUGGAAUGCAUCUAAUGGCAUGGCAUGGCAUAUUACACAUACGCCCCGUGGCGCAUUGUCUAAUAAAUACAAAU-UUAGACAGCAUGAUUUGCCAGAGCAACGUCCGCCUCGUAGCCAGA ((((..(((......(((((...((((..............((((....))))..((((((((...........-))))))))))))...)))))((((.......).)))))).)))). ( -33.00) >consensus UUGGAAUGCAUCUAAUGGCAUAGCAUGGCAUAUUACACAUACGCCCCGUGGCGCAUUGUCUAAUAAAUACAAAU_UUAGACAGCAUGAUUUGCCAGC_______________________ ...............(((((...((((..............((((....))))..((((((((............))))))))))))...)))))......................... (-18.45 = -19.37 + 0.92)

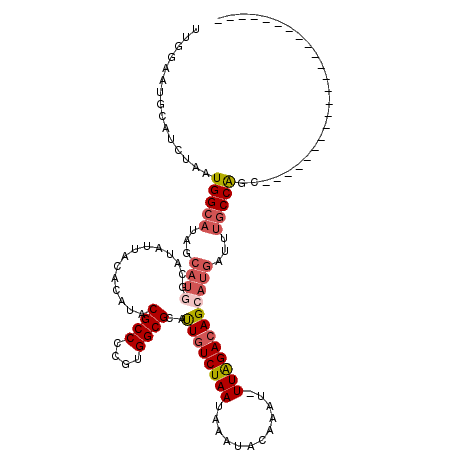
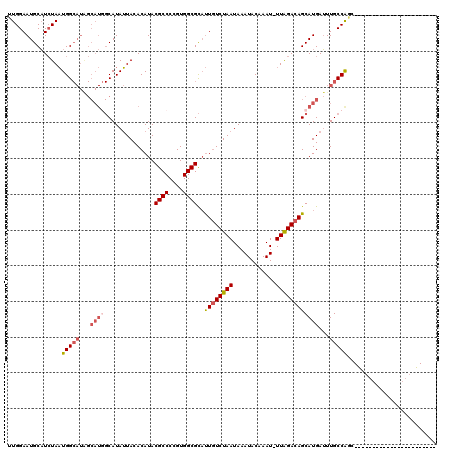
| Location | 12,364,928 – 12,365,021 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 96.70 |
| Mean single sequence MFE | -22.53 |
| Consensus MFE | -20.30 |
| Energy contribution | -21.13 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.577387 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12364928 93 - 27905053 GCUGUCAUGUUGGAUUGGAAUGCAUCUAAUGGCAUAGCAUGGCAUAUUACACAUACGCCCCGUGGCGCAUUGUCUAAUAAAUACAAAUUUAGA ..((((((((((.((((((.....))))))....))))))))))(((((.(((..((((....))))...))).))))).............. ( -24.10) >DroPse_CAF1 32010 93 - 1 GCUGUCAUGUUGGACUGGAAUGCAUCUAAUGGCAUGGCAUGGCAUAUUACACAUACGCCCCGUGGCGCAUUGUCUAAUAAAUACAAAUUUAGA ((((((((((((...((((.....)))).)))))))))..))).(((((.(((..((((....))))...))).))))).............. ( -22.90) >DroSec_CAF1 33363 93 - 1 GCUGUCAUGUUGGAUUGGACUGCAUCUAAUGGCAUAGCAUGGCAUAUUACACAUACGCCCCGUGGCGCAUUGUCUAAUAAAUACAAAUUUAGA ..((((((((((.((((((.....))))))....))))))))))(((((.(((..((((....))))...))).))))).............. ( -23.90) >DroYak_CAF1 29871 93 - 1 GCUGUCAUGUUGGAUUGGAAUGCAUCUAAUGGCAUAGCAUUGCAUAUUACACAUACGCCCCGUGGCGCAUUUUCUAAUAAAUACAAAUUUAGA ..(((..(((((((...((((((...((((((((......))).))))).......(((....))))))))))))))))...)))........ ( -19.60) >DroAna_CAF1 15733 93 - 1 GCUGUCAUGUCGGUUUGGAAUGCAUCUAAUGGCAUAGCAUGGCAUAUUACACAUACGCCCUGUGGCGCAUUGUCUAAUAAAUACAAAUUUAGA ..((((((((..(((((((.....)))))..))...))))))))(((((.(((..((((....))))...))).))))).............. ( -21.80) >DroPer_CAF1 29111 93 - 1 GCUGUCAUGUUGGACUGGAAUGCAUCUAAUGGCAUGGCAUGGCAUAUUACACAUACGCCCCGUGGCGCAUUGUCUAAUAAAUACAAAUUUAGA ((((((((((((...((((.....)))).)))))))))..))).(((((.(((..((((....))))...))).))))).............. ( -22.90) >consensus GCUGUCAUGUUGGAUUGGAAUGCAUCUAAUGGCAUAGCAUGGCAUAUUACACAUACGCCCCGUGGCGCAUUGUCUAAUAAAUACAAAUUUAGA ..((((((((((.((((((.....))))))....))))))))))(((((.(((..((((....))))...))).))))).............. (-20.30 = -21.13 + 0.83)

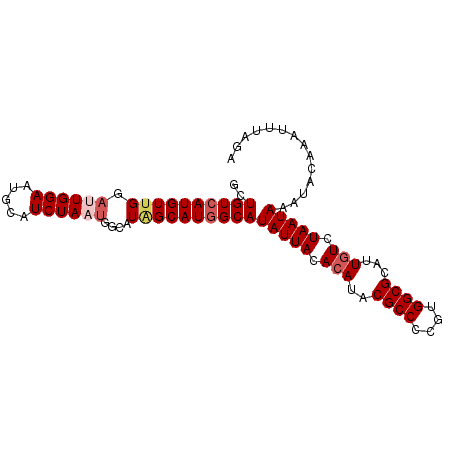
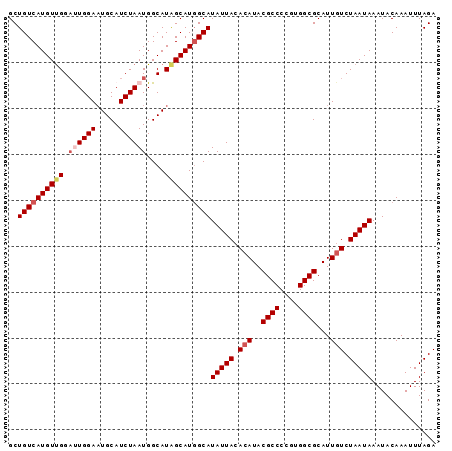
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:02 2006