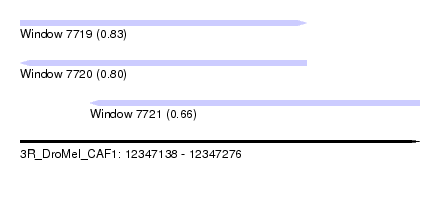
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,347,138 – 12,347,276 |
| Length | 138 |
| Max. P | 0.827971 |

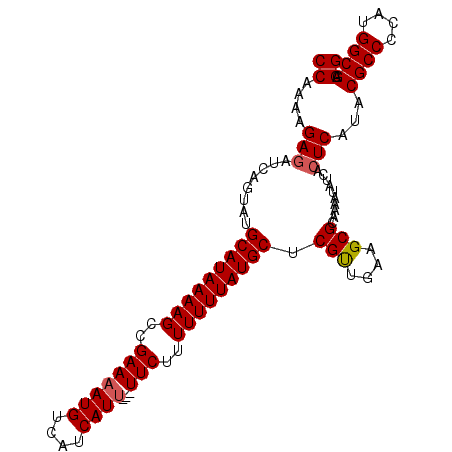
| Location | 12,347,138 – 12,347,237 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 96.01 |
| Mean single sequence MFE | -24.54 |
| Consensus MFE | -19.08 |
| Energy contribution | -19.32 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827971 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12347138 99 + 27905053 CCAAAAGAGAUCUGUAUGCAUAAAAGCCGAAAAUGUCAUCAUU---UUCUUUUUUUAUGCCCGUUGAAGCGCAAAAUAUCACUCAUACGCCCCAUGGCGAGG ((....(((........(((((((((..(((((((....))))---)))..))))))))).(((....)))..........)))...((((....)))).)) ( -24.90) >DroSec_CAF1 15911 99 + 1 CCAAAAGAGAUCAGUAUGCAUAAAAGCCGAAAAUGUCAUCAUU---UUCUUUUUUUAUGCUCGUUGAAGCGCAAAAUAUCACUCAUACGCCCCAUGGCGAGG ((....(((........(((((((((..(((((((....))))---)))..))))))))).(((....)))..........)))...((((....)))).)) ( -25.10) >DroSim_CAF1 15041 99 + 1 CCAAAAGAGAUCAGUAUGCAUAAAAGCCGAAAAUGUCAUCAUU---UUCUUUUUUUAUGCUCGUUGAAGCGCAAAAUAUCACUCAUACGCCCCAUGGCGAGG ((....(((........(((((((((..(((((((....))))---)))..))))))))).(((....)))..........)))...((((....)))).)) ( -25.10) >DroEre_CAF1 15809 99 + 1 CCAAAAGAGAUCUGUAUGCAUAAAAGCCGAAAAUGUCAUCAUU---UUCUUUUUUUAUGCUCGUUGAAGCGCAAAAUAUCACUCAUACGCCCCAUGGCGAGG ((....(((........(((((((((..(((((((....))))---)))..))))))))).(((....)))..........)))...((((....)))).)) ( -25.10) >DroYak_CAF1 11913 102 + 1 CCAAACGAUAUCAGUAUGCAUAAAAGCCGAAAAUGUCAUCAUCAAAUUCUUUUUUUAUGCCCGCUGAAGCGCAAAAUAUCACUCAUACGCCCCAUGGCGAGG ((...((...(((((..(((((((((..(((..((.......))..)))..)))))))))..)))))..))................((((....)))).)) ( -22.50) >consensus CCAAAAGAGAUCAGUAUGCAUAAAAGCCGAAAAUGUCAUCAUU___UUCUUUUUUUAUGCUCGUUGAAGCGCAAAAUAUCACUCAUACGCCCCAUGGCGAGG ((....(((........(((((((((..(((((((....))))...)))..))))))))).(((....)))..........)))...((((....)))).)) (-19.08 = -19.32 + 0.24)

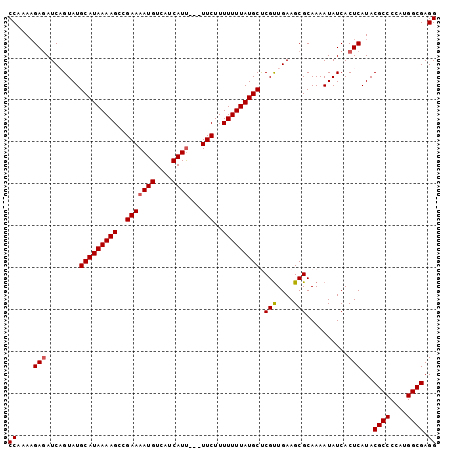
| Location | 12,347,138 – 12,347,237 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 96.01 |
| Mean single sequence MFE | -27.54 |
| Consensus MFE | -22.46 |
| Energy contribution | -22.06 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.802515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12347138 99 - 27905053 CCUCGCCAUGGGGCGUAUGAGUGAUAUUUUGCGCUUCAACGGGCAUAAAAAAAGAA---AAUGAUGACAUUUUCGGCUUUUAUGCAUACAGAUCUCUUUUGG .....((.(((((((((.((......)).)))))))))..))((((((((...(((---((((....)))))))...))))))))...((((.....)))). ( -28.40) >DroSec_CAF1 15911 99 - 1 CCUCGCCAUGGGGCGUAUGAGUGAUAUUUUGCGCUUCAACGAGCAUAAAAAAAGAA---AAUGAUGACAUUUUCGGCUUUUAUGCAUACUGAUCUCUUUUGG .....((((((((((((.((......)).)))))))))....((((((((...(((---((((....)))))))...))))))))..............))) ( -27.40) >DroSim_CAF1 15041 99 - 1 CCUCGCCAUGGGGCGUAUGAGUGAUAUUUUGCGCUUCAACGAGCAUAAAAAAAGAA---AAUGAUGACAUUUUCGGCUUUUAUGCAUACUGAUCUCUUUUGG .....((((((((((((.((......)).)))))))))....((((((((...(((---((((....)))))))...))))))))..............))) ( -27.40) >DroEre_CAF1 15809 99 - 1 CCUCGCCAUGGGGCGUAUGAGUGAUAUUUUGCGCUUCAACGAGCAUAAAAAAAGAA---AAUGAUGACAUUUUCGGCUUUUAUGCAUACAGAUCUCUUUUGG ..(((...(((((((((.((......)).))))))))).)))((((((((...(((---((((....)))))))...))))))))...((((.....)))). ( -28.60) >DroYak_CAF1 11913 102 - 1 CCUCGCCAUGGGGCGUAUGAGUGAUAUUUUGCGCUUCAGCGGGCAUAAAAAAAGAAUUUGAUGAUGACAUUUUCGGCUUUUAUGCAUACUGAUAUCGUUUGG ..((((..(((((((((.((......)).)))))))))))))((((((((...(((..((.......))..)))...))))))))................. ( -25.90) >consensus CCUCGCCAUGGGGCGUAUGAGUGAUAUUUUGCGCUUCAACGAGCAUAAAAAAAGAA___AAUGAUGACAUUUUCGGCUUUUAUGCAUACUGAUCUCUUUUGG ..(((...(((((((((.((......)).))))))))).)))((((((((...(((...((((....)))))))...))))))))................. (-22.46 = -22.06 + -0.40)

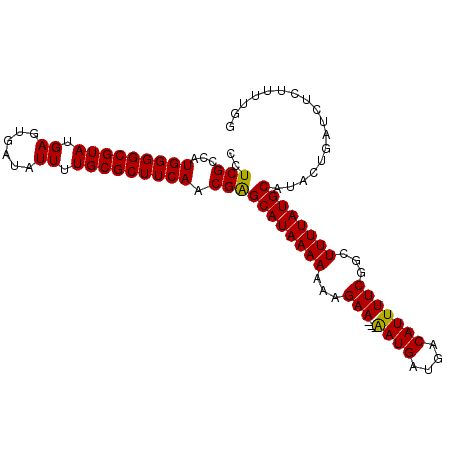
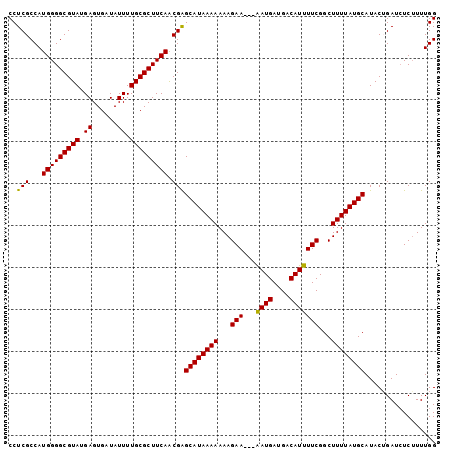
| Location | 12,347,162 – 12,347,276 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.87 |
| Mean single sequence MFE | -34.38 |
| Consensus MFE | -21.54 |
| Energy contribution | -23.40 |
| Covariance contribution | 1.86 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.664020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12347162 114 - 27905053 GCGUGGAAAUCUCUUGCGGCCUAAUGAGCCGUACUUGGG---CCUCGCCAUGGGGCGUAUGAGUGAUAUUUUGCGCUUCAACGGGCAUAAAAAAAGAA---AAUGAUGACAUUUUCGGCU ..............(((((((....).))))))....((---((..(((.(((((((((.((......)).)))))))))...))).........(((---((((....))))))))))) ( -37.20) >DroPse_CAF1 17062 92 - 1 GAG--------CCA--UGGCCUAAUGAGCCGUACUUGGGAGCCAGCGCCAUGGGGCGUAUGGGUGAUAUUUUUCGCUUCAGCGGGCAACAAAU--AAA---AAUGA-------------U ..(--------(((--(((((....).)))))..(((((..((((((((....))))).)))((((......)))))))))..))).......--...---.....-------------. ( -26.50) >DroSec_CAF1 15935 114 - 1 GCGUGGAAAUCUCAUGCGGCCUAAUGAGCCGUACUUGGG---CCUCGCCAUGGGGCGUAUGAGUGAUAUUUUGCGCUUCAACGAGCAUAAAAAAAGAA---AAUGAUGACAUUUUCGGCU ((((((.....))))))(((((((..........)))))---))..(((.(((((((((.((......)).)))))))))...............(((---((((....)))))))))). ( -38.40) >DroSim_CAF1 15065 114 - 1 GCGUGGAAAUCUCAUGCGGCCUAAUGAGCCGUACUUGGG---CCUCGCCAUGGGGCGUAUGAGUGAUAUUUUGCGCUUCAACGAGCAUAAAAAAAGAA---AAUGAUGACAUUUUCGGCU ((((((.....))))))(((((((..........)))))---))..(((.(((((((((.((......)).)))))))))...............(((---((((....)))))))))). ( -38.40) >DroYak_CAF1 11937 117 - 1 GCGUGGAAAUCUCUUGCGGCCUAAUGAGCCGUACUCGGG---CCUCGCCAUGGGGCGUAUGAGUGAUAUUUUGCGCUUCAGCGGGCAUAAAAAAAGAAUUUGAUGAUGACAUUUUCGGCU ((.(((((((.((.(((((((....).)))))).....(---((.(((..(((((((((.((......)).))))))))))))))).....................)).))))))))). ( -37.20) >DroPer_CAF1 12877 92 - 1 GAG--------CCA--UGGCCUAAUGAGCCAUACUUGGGAGCCCACGCCAUGGGGCGUAUGGGUGAUAUUUUUCGCUUCAGCGGGCAACAAAU--AAA---AAUGA-------------U ...--------...--..((((..((((((((((.(((....))).(((....)))))))))((((......))))))))..)))).......--...---.....-------------. ( -28.60) >consensus GCGUGGAAAUCUCAUGCGGCCUAAUGAGCCGUACUUGGG___CCUCGCCAUGGGGCGUAUGAGUGAUAUUUUGCGCUUCAACGGGCAUAAAAAAAGAA___AAUGAUGACAUUUUCGGCU ..(.((....(((((((((((....).))))))..))))...)).)(((.(((((((((.((......)).)))))))))...))).................................. (-21.54 = -23.40 + 1.86)

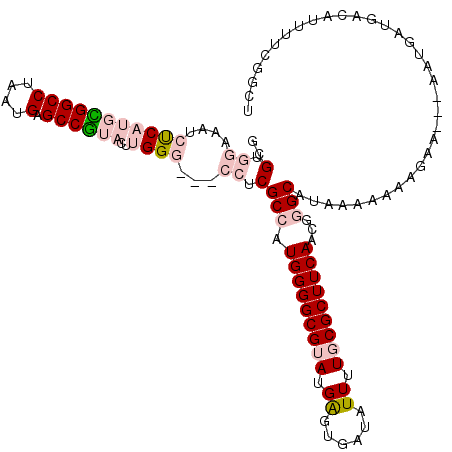
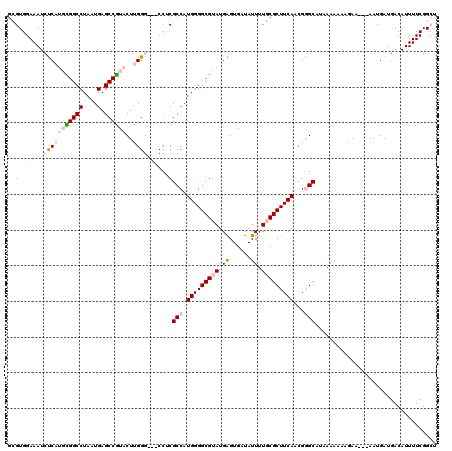
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:50 2006