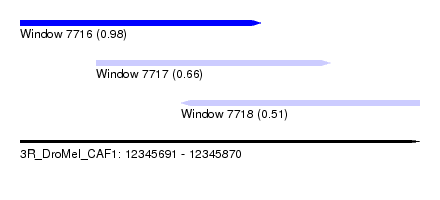
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,345,691 – 12,345,870 |
| Length | 179 |
| Max. P | 0.981153 |

| Location | 12,345,691 – 12,345,799 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 86.02 |
| Mean single sequence MFE | -34.56 |
| Consensus MFE | -28.40 |
| Energy contribution | -28.84 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981153 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
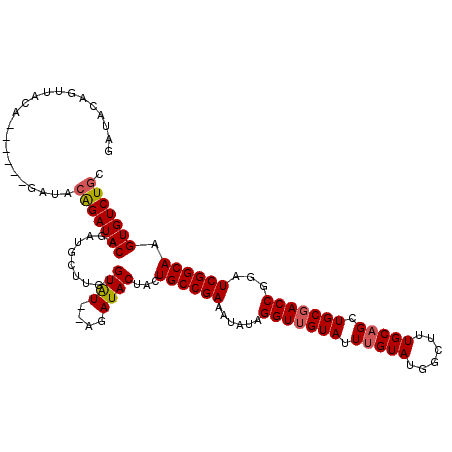
>3R_DroMel_CAF1 12345691 108 + 27905053 GAUACAGUUACA------GAUACAGAUACAGAUGCUUGUGUAU--AGAUACUACUGCCGAAAUAUAGGUUGUAUUUGUAUGGCUUUGCAGCUGCGACCGGAUCGGCAAGUGUCUGC ..........((------(((((..(((((.(....).)))))--.........((((((......(((((((.(((((......))))).)))))))...)))))).))))))). ( -35.00) >DroSec_CAF1 14485 114 + 1 GAUACAGUUACAGAUACAGAUACAGAUACAGAUGCUUCUGUAU--AGAUACUACUGCCGAAAUAUAGGUUGUAUUUGUAUGGCUUUGCAGCUGCGACCGGAUCGGCAAGUGUCUGC ................(((((((..(((((((....)))))))--.........((((((......(((((((.(((((......))))).)))))))...)))))).))))))). ( -39.10) >DroSim_CAF1 13615 114 + 1 GAUACAGUUACAGUUACAGAUACAGAUACAGAUGCUUGUGUAU--AGAUACCACUGCCGAAAUAUAGGUUGUAUUUGUAUGGCUUUGCAGCUGCGACCGGAUCGGCAAGUGUCUGC ................(((((((..(((((.(....).)))))--.........((((((......(((((((.(((((......))))).)))))))...)))))).))))))). ( -35.00) >DroEre_CAF1 14412 91 + 1 GAUACAGAUUCA------AAUAGGGAUACAGAUGC-------------------UGCCGAAAUAUAGGUUGUAUUUGUAUGGCUUUGCUGCUGCGACCGGAUCGGCAAGUGUCUGC .......((((.------.....)))).(((((((-------------------((((((......(((((((...(((......)))...)))))))...))))).)))))))). ( -28.90) >DroYak_CAF1 10421 110 + 1 GAUACGGAUACA------GAUACGGAUACAGAUGCUUGUGUGUAGAGAUACUACUGCCGAAAUAUAGGUGGUAUUUGUAUGGCUUUGCAGCUGCGACCGGAUCGGCAAGUGUCUGC ..........((------(((((..((((((....))))))((((.....))))((((((......(((.(((.(((((......))))).))).)))...)))))).))))))). ( -34.80) >consensus GAUACAGUUACA______GAUACAGAUACAGAUGCUUGUGUAU__AGAUACUACUGCCGAAAUAUAGGUUGUAUUUGUAUGGCUUUGCAGCUGCGACCGGAUCGGCAAGUGUCUGC ......................(((((((..........((((....))))...((((((......(((((((.(((((......))))).)))))))...)))))).))))))). (-28.40 = -28.84 + 0.44)

| Location | 12,345,725 – 12,345,830 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 89.55 |
| Mean single sequence MFE | -33.04 |
| Consensus MFE | -25.92 |
| Energy contribution | -28.40 |
| Covariance contribution | 2.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.663581 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
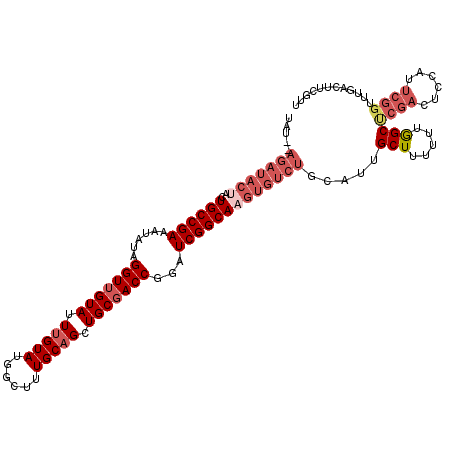
>3R_DroMel_CAF1 12345725 105 + 27905053 UAU--AGAUACUACUGCCGAAAUAUAGGUUGUAUUUGUAUGGCUUUGCAGCUGCGACCGGAUCGGCAAGUGUCUGCAUUGCUUUUUUGGCUCGACU---------UUGACUUCGUU ..(--(((((((..((((((......(((((((.(((((......))))).)))))))...))))))))))))))....(((.....))).(((..---------......))).. ( -32.70) >DroSec_CAF1 14525 114 + 1 UAU--AGAUACUACUGCCGAAAUAUAGGUUGUAUUUGUAUGGCUUUGCAGCUGCGACCGGAUCGGCAAGUGUCUGCAUUGCUUUUUUGGCCCGACUCCAUUCGGUUUGACUUCGUU ..(--(((((((..((((((......(((((((.(((((......))))).)))))))...))))))))))))))....(((.....)))((((......))))............ ( -36.00) >DroSim_CAF1 13655 114 + 1 UAU--AGAUACCACUGCCGAAAUAUAGGUUGUAUUUGUAUGGCUUUGCAGCUGCGACCGGAUCGGCAAGUGUCUGCAUUGCUUUUUUGGCCCGACUCCAUUCGGUUUGACUUCGUU ...--.((...((..((((((.....(((((((.(((((......))))).)))))))(((((((..((((....))))(((.....))))))).))).)))))).))...))... ( -34.80) >DroEre_CAF1 14441 102 + 1 --------------UGCCGAAAUAUAGGUUGUAUUUGUAUGGCUUUGCUGCUGCGACCGGAUCGGCAAGUGUCUGCAUUGCUUUUUUGGCUCGACUCCAUUCGGUUUGACUGCGUU --------------.((((((.....(((((((...(((......)))...)))))))(((((((((......)))...(((.....))).))).))).))))))........... ( -28.90) >DroYak_CAF1 10455 116 + 1 UGUAGAGAUACUACUGCCGAAAUAUAGGUGGUAUUUGUAUGGCUUUGCAGCUGCGACCGGAUCGGCAAGUGUCUGCAUUGCUUUUUUAGCUCGACUCCAUUCGGUUUGACUUCGUU (((..(((((((..((((((......(((.(((.(((((......))))).))).)))...))))))))))))))))..(((.....)))((((..((....)).))))....... ( -32.80) >consensus UAU__AGAUACUACUGCCGAAAUAUAGGUUGUAUUUGUAUGGCUUUGCAGCUGCGACCGGAUCGGCAAGUGUCUGCAUUGCUUUUUUGGCUCGACUCCAUUCGGUUUGACUUCGUU .....(((((((..((((((......(((((((.(((((......))))).)))))))...))))))))))))).....(((.....)))((((......))))............ (-25.92 = -28.40 + 2.48)

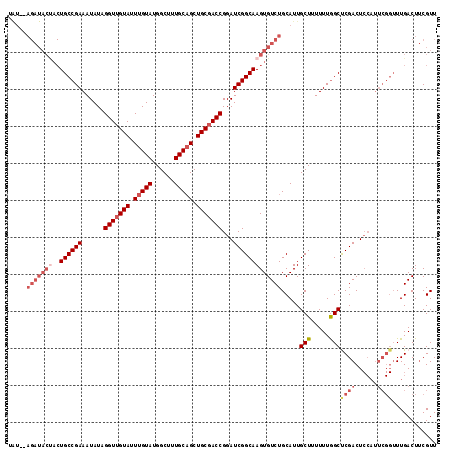
| Location | 12,345,763 – 12,345,870 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.61 |
| Mean single sequence MFE | -29.63 |
| Consensus MFE | -20.64 |
| Energy contribution | -21.75 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.510770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
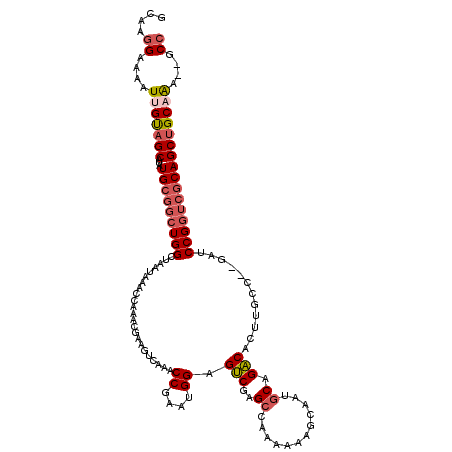
>3R_DroMel_CAF1 12345763 107 - 27905053 GCAAGGAAAAUUGUAGCAUAUGCGGCUGGCUAAUAAACCAAACGAAGUCAA---------AGUCGAGCCAAAAAAGCAAUGCAGACACUUGCC--GAUCCGGUCGCAGCUGCAAA--GCC ....((....(((((((...(((((((((..............(....)..---------.((((.((.......))...((((....)))))--))))))))))))))))))).--.)) ( -27.40) >DroSec_CAF1 14563 116 - 1 GCAAGGAAAAUUGUAGCAUAUGCGGCUGGCUAAUAAACCAAACGAAGUCAAACCGAAUGGAGUCGGGCCAAAAAAGCAAUGCAGACACUUGCC--GAUCCGGUCGCAGCUGCAAA--GCC ....((....(((((((...(((((((((........(((..((.........))..))).((((.((.......))...((((....)))))--))))))))))))))))))).--.)) ( -31.30) >DroSim_CAF1 13693 116 - 1 GCAAGGAAAAUUGUAGCAUAUGCGGCUGGCUAAUAAACCAAACGAAGUCAAACCGAAUGGAGUCGGGCCAAAAAAGCAAUGCAGACACUUGCC--GAUCCGGUCGCAGCUGCAAA--GCC ....((....(((((((...(((((((((........(((..((.........))..))).((((.((.......))...((((....)))))--))))))))))))))))))).--.)) ( -31.30) >DroEre_CAF1 14467 116 - 1 GCAAGGAAAAUCGUAGCAUAUGCGGCUGGCUAAUAAACCAAACGCAGUCAAACCGAAUGGAGUCGAGCCAAAAAAGCAAUGCAGACACUUGCC--GAUCCGGUCGCAGCAGCAAA--GCC ............((.((...((((..(((........)))..)))).......(((.((((.(((.((.......))...((((....)))))--)))))).)))..)).))...--... ( -25.50) >DroYak_CAF1 10495 116 - 1 GCAAGGAAAAUUGUAGCAUAUGCGGCUGGCUAAUAAACCAAACGAAGUCAAACCGAAUGGAGUCGAGCUAAAAAAGCAAUGCAGACACUUGCC--GAUCCGGUCGCAGCUGCAAA--GCC ....((....(((((((...(((((((((........(((..((.........))..))).((((.(((.....)))...((((....)))))--))))))))))))))))))).--.)) ( -33.20) >DroPer_CAF1 11540 91 - 1 GGAGAGAAAAUCGCAGCAUAUG----UGGCUAAUAAACCAGACCAAA--------------GCCCAG-----------GCCCAGGCACUUGCAGGGAGCCGGCCACAGCUGCAGUCAGCG ............(((((...((----(((((.........(......--------------...).(-----------((((..((....))..)).)))))))))))))))........ ( -29.10) >consensus GCAAGGAAAAUUGUAGCAUAUGCGGCUGGCUAAUAAACCAAACGAAGUCAAACCGAAUGGAGUCGAGCCAAAAAAGCAAUGCAGACACUUGCC__GAUCCGGUCGCAGCUGCAAA__GCC ....((....(((((((...(((((((((.......................((....)).(((..((............)).)))............))))))))))))))))....)) (-20.64 = -21.75 + 1.11)

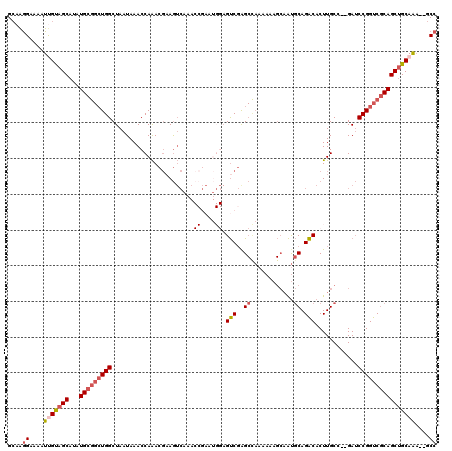
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:47 2006