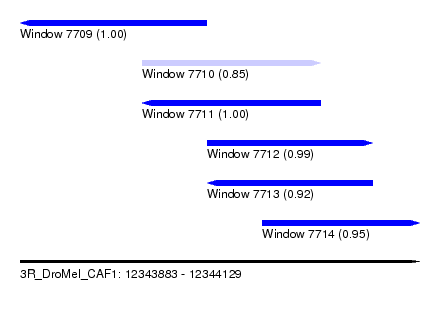
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,343,883 – 12,344,129 |
| Length | 246 |
| Max. P | 0.999507 |

| Location | 12,343,883 – 12,343,998 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 99.65 |
| Mean single sequence MFE | -33.86 |
| Consensus MFE | -33.00 |
| Energy contribution | -33.20 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.67 |
| SVM RNA-class probability | 0.999507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12343883 115 - 27905053 UUUUUUCUCACUAAUUAAUAAAUGUUGCUGUUGUUGGCGCUGGUUUCGUUUGCCGCUGUGUUGCUGAUAUCUUUUGUGCAGCGAAAUGGUUUAUUGGCCUAUUAGCGCAGCAUUU ...................(((((((((.((((.(((.((..((.((((((..(((((..(....(....)....)..)))))))))))...))..))))).))))))))))))) ( -33.70) >DroSec_CAF1 12630 115 - 1 UUUUUUCUCACUAAUUAAUAAAUGUUGCUGUUGUUGGCGCUGGUUUCGGUUGCCGCUGUGUUGCUGAUAUCUUUUGUGCAGCGAAAUGGUUUAUUGGCCUAUUAGCGCAGCAUUU ...................(((((((((.((((.(((.((..((..(.(((..(((((..(....(....)....)..))))).))).)...))..))))).))))))))))))) ( -33.90) >DroSim_CAF1 11763 115 - 1 UUUUUUCUCACUAAUUAAUAAAUGUUGCUGUUGUUGGCGCUGGUUUCGGUUGCCGCUGUGUUGCUGAUAUCUUUUGUGCAGCGAAAUGGUUUAUUGGCCUAUUAGCGCAGCAUUU ...................(((((((((.((((.(((.((..((..(.(((..(((((..(....(....)....)..))))).))).)...))..))))).))))))))))))) ( -33.90) >DroEre_CAF1 12594 115 - 1 UUUUUUCUCACUAAUUAAUAAAUGUUGCUGUUGUUGGCGCUGGUUUCGGUUGCCGCUGUGUUGCUGAUAUCUUUUGUGCAGCGAAAUGGUUUAUUGGCCUAUUAGCGCAGCAUUU ...................(((((((((.((((.(((.((..((..(.(((..(((((..(....(....)....)..))))).))).)...))..))))).))))))))))))) ( -33.90) >DroYak_CAF1 8573 115 - 1 UUUUUUCUCACUAAUUAAUAAAUGUUGCUGUUGUUGGCGCUGGUUUCGGUUGCCGCUGUGUUGCUGAUAUCUUUUGUGCAGCGAAAUGGUUUAUUGGCCUAUUAGCGCAGCAUUU ...................(((((((((.((((.(((.((..((..(.(((..(((((..(....(....)....)..))))).))).)...))..))))).))))))))))))) ( -33.90) >consensus UUUUUUCUCACUAAUUAAUAAAUGUUGCUGUUGUUGGCGCUGGUUUCGGUUGCCGCUGUGUUGCUGAUAUCUUUUGUGCAGCGAAAUGGUUUAUUGGCCUAUUAGCGCAGCAUUU ...................(((((((((.((((.(((.((..((..(.(((..(((((..(....(....)....)..))))).))).)...))..))))).))))))))))))) (-33.00 = -33.20 + 0.20)



| Location | 12,343,958 – 12,344,068 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 98.73 |
| Mean single sequence MFE | -28.29 |
| Consensus MFE | -26.65 |
| Energy contribution | -27.05 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.850073 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12343958 110 + 27905053 GCGCCAACAACAGCAACAUUUAUUAAUUAGUGAGAAAAAAUUAAUGCGCAACUGUCAUCAUUACUCGUGCCCCAUCCACAGGAUCUGUGGACAUUGGUGGCGGAUAUAGU ((((............((((........)))).............)))).(((((.(((........((((((((((((((...)))))))...))).)))))))))))) ( -25.81) >DroSec_CAF1 12705 110 + 1 GCGCCAACAACAGCAACAUUUAUUAAUUAGUGAGAAAAAAUUAAUGCGCAACUGUCAUCAUUACUCCUGCCCCAUCCACAGGAUCUGUGGACAUUGGUGGCGGAUAUGGU ((((............((((........)))).............)))).......(((((...(((.(((((((((((((...)))))))...))).)))))).))))) ( -28.91) >DroSim_CAF1 11838 110 + 1 GCGCCAACAACAGCAACAUUUAUUAAUUAGUGAGAAAAAAUUAAUGCGCAACUGUCAUCAUUACUCCUGCCCCAUCCACAGGAUCUGUGGACAUUGGUGGCGGAUAUGGU ((((............((((........)))).............)))).......(((((...(((.(((((((((((((...)))))))...))).)))))).))))) ( -28.91) >DroEre_CAF1 12669 110 + 1 GCGCCAACAACAGCAACAUUUAUUAAUUAGUGAGAAAAAAUUAAUGCGCAACUGUCAUCAUUACUCCUGCCCCAUCCACAGGAUCUGUGGACAUUGGCGGCGGAUAUGGU ((((............((((........)))).............)))).......(((((...(((.(((((((((((((...)))))))...))).)))))).))))) ( -28.91) >DroYak_CAF1 8648 110 + 1 GCGCCAACAACAGCAACAUUUAUUAAUUAGUGAGAAAAAAUUAAUGCGCAACUGUCAUCAUUACUCCUGCCCCAUCCACAGGAUCUGUGGACAUUGGCGGCGGAUAUGGU ((((............((((........)))).............)))).......(((((...(((.(((((((((((((...)))))))...))).)))))).))))) ( -28.91) >consensus GCGCCAACAACAGCAACAUUUAUUAAUUAGUGAGAAAAAAUUAAUGCGCAACUGUCAUCAUUACUCCUGCCCCAUCCACAGGAUCUGUGGACAUUGGUGGCGGAUAUGGU ((((............((((........)))).............)))).......(((((...(((.(((((((((((((...)))))))...))).)))))).))))) (-26.65 = -27.05 + 0.40)

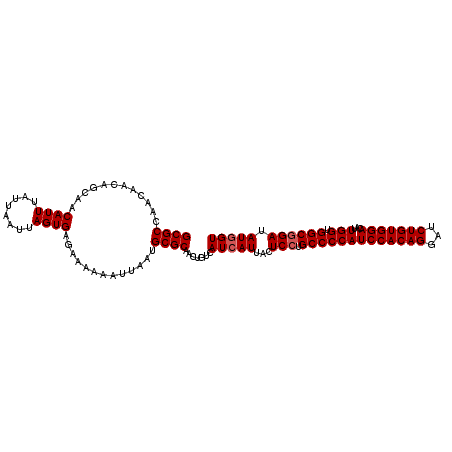

| Location | 12,343,958 – 12,344,068 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 98.73 |
| Mean single sequence MFE | -33.90 |
| Consensus MFE | -32.16 |
| Energy contribution | -32.36 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.10 |
| SVM RNA-class probability | 0.998428 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12343958 110 - 27905053 ACUAUAUCCGCCACCAAUGUCCACAGAUCCUGUGGAUGGGGCACGAGUAAUGAUGACAGUUGCGCAUUAAUUUUUUCUCACUAAUUAAUAAAUGUUGCUGUUGUUGGCGC ........((((.((...((((((((...)))))))).))...........((..(((((.(((.(((((((..........)))))))...))).)))))..)))))). ( -31.30) >DroSec_CAF1 12705 110 - 1 ACCAUAUCCGCCACCAAUGUCCACAGAUCCUGUGGAUGGGGCAGGAGUAAUGAUGACAGUUGCGCAUUAAUUUUUUCUCACUAAUUAAUAAAUGUUGCUGUUGUUGGCGC .((...((((((.(((...(((((((...))))))))))))).))).....((..(((((.(((.(((((((..........)))))))...))).)))))..))))... ( -33.80) >DroSim_CAF1 11838 110 - 1 ACCAUAUCCGCCACCAAUGUCCACAGAUCCUGUGGAUGGGGCAGGAGUAAUGAUGACAGUUGCGCAUUAAUUUUUUCUCACUAAUUAAUAAAUGUUGCUGUUGUUGGCGC .((...((((((.(((...(((((((...))))))))))))).))).....((..(((((.(((.(((((((..........)))))))...))).)))))..))))... ( -33.80) >DroEre_CAF1 12669 110 - 1 ACCAUAUCCGCCGCCAAUGUCCACAGAUCCUGUGGAUGGGGCAGGAGUAAUGAUGACAGUUGCGCAUUAAUUUUUUCUCACUAAUUAAUAAAUGUUGCUGUUGUUGGCGC ........(((((((...((((((((...))))))))..))).........((..(((((.(((.(((((((..........)))))))...))).)))))..)))))). ( -35.30) >DroYak_CAF1 8648 110 - 1 ACCAUAUCCGCCGCCAAUGUCCACAGAUCCUGUGGAUGGGGCAGGAGUAAUGAUGACAGUUGCGCAUUAAUUUUUUCUCACUAAUUAAUAAAUGUUGCUGUUGUUGGCGC ........(((((((...((((((((...))))))))..))).........((..(((((.(((.(((((((..........)))))))...))).)))))..)))))). ( -35.30) >consensus ACCAUAUCCGCCACCAAUGUCCACAGAUCCUGUGGAUGGGGCAGGAGUAAUGAUGACAGUUGCGCAUUAAUUUUUUCUCACUAAUUAAUAAAUGUUGCUGUUGUUGGCGC ......((((((.(((...(((((((...))))))))))))).)))((...((..(((((.(((.(((((((..........)))))))...))).)))))..)).)).. (-32.16 = -32.36 + 0.20)


| Location | 12,343,998 – 12,344,100 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 91.05 |
| Mean single sequence MFE | -33.23 |
| Consensus MFE | -30.67 |
| Energy contribution | -31.17 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.36 |
| SVM RNA-class probability | 0.992961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12343998 102 + 27905053 UUAAUGCGCAACUGUCAUCAUUACUCGUGCCCCAUCCACAGGAUCUGUGGACAUUGGUGGCGGAUAUAGUUUGUUGUGUGUGUUCUUCCUCACACACCA-CCC---- .....(((.((((((.(((........((((((((((((((...)))))))...))).)))))))))))))))).(((((((........)))))))..-...---- ( -31.50) >DroSec_CAF1 12745 106 + 1 UUAAUGCGCAACUGUCAUCAUUACUCCUGCCCCAUCCACAGGAUCUGUGGACAUUGGUGGCGGAUAUGGUUUGUUGUGUGUGUUCUUCCCCACACCCCA-CCCACCG .....(((((((....(((((...(((.(((((((((((((...)))))))...))).)))))).)))))..)))))))((((........))))....-....... ( -33.70) >DroSim_CAF1 11878 106 + 1 UUAAUGCGCAACUGUCAUCAUUACUCCUGCCCCAUCCACAGGAUCUGUGGACAUUGGUGGCGGAUAUGGUUUGUUGUGUGUGUUCUUCCCCACACCCCA-CCCCCCG .....(((((((....(((((...(((.(((((((((((((...)))))))...))).)))))).)))))..)))))))((((........))))....-....... ( -33.70) >DroEre_CAF1 12709 103 + 1 UUAAUGCGCAACUGUCAUCAUUACUCCUGCCCCAUCCACAGGAUCUGUGGACAUUGGCGGCGGAUAUGGUUUGUUGUGUGUGUUU-UCCGCACCCCCCAAUA---AA .....(((((((....(((((...(((.(((((((((((((...)))))))...))).)))))).)))))..)))))))((((..-...)))).........---.. ( -34.00) >consensus UUAAUGCGCAACUGUCAUCAUUACUCCUGCCCCAUCCACAGGAUCUGUGGACAUUGGUGGCGGAUAUGGUUUGUUGUGUGUGUUCUUCCCCACACCCCA_CCC__CG .....(((((((....(((((...(((.(((((((((((((...)))))))...))).)))))).)))))..)))))))(((........))).............. (-30.67 = -31.17 + 0.50)


| Location | 12,343,998 – 12,344,100 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 91.05 |
| Mean single sequence MFE | -34.70 |
| Consensus MFE | -28.95 |
| Energy contribution | -29.45 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.920771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12343998 102 - 27905053 ----GGG-UGGUGUGUGAGGAAGAACACACACAACAAACUAUAUCCGCCACCAAUGUCCACAGAUCCUGUGGAUGGGGCACGAGUAAUGAUGACAGUUGCGCAUUAA ----(((-(((((((((........))))))).........)))))(((.(((...(((((((...)))))))))))))..(.(((((.......))))).)..... ( -32.70) >DroSec_CAF1 12745 106 - 1 CGGUGGG-UGGGGUGUGGGGAAGAACACACACAACAAACCAUAUCCGCCACCAAUGUCCACAGAUCCUGUGGAUGGGGCAGGAGUAAUGAUGACAGUUGCGCAUUAA .(((..(-(...(((((........)))))...))..)))...((((((.(((...(((((((...))))))))))))).)))(((((.......)))))....... ( -38.00) >DroSim_CAF1 11878 106 - 1 CGGGGGG-UGGGGUGUGGGGAAGAACACACACAACAAACCAUAUCCGCCACCAAUGUCCACAGAUCCUGUGGAUGGGGCAGGAGUAAUGAUGACAGUUGCGCAUUAA ...((..-((..(((((.(......).)))))..))..))...((((((.(((...(((((((...))))))))))))).)))(((((.......)))))....... ( -38.40) >DroEre_CAF1 12709 103 - 1 UU---UAUUGGGGGGUGCGGA-AAACACACACAACAAACCAUAUCCGCCGCCAAUGUCCACAGAUCCUGUGGAUGGGGCAGGAGUAAUGAUGACAGUUGCGCAUUAA ..---..(((..(.(((.(..-...).))).)..)))......((((((.(((...(((((((...))))))))))))).)))(((((.......)))))....... ( -29.70) >consensus CG__GGG_UGGGGUGUGGGGAAGAACACACACAACAAACCAUAUCCGCCACCAAUGUCCACAGAUCCUGUGGAUGGGGCAGGAGUAAUGAUGACAGUUGCGCAUUAA ........((..(((((.(......).)))))..)).......((((((.(((...(((((((...))))))))))))).)))(((((.......)))))....... (-28.95 = -29.45 + 0.50)

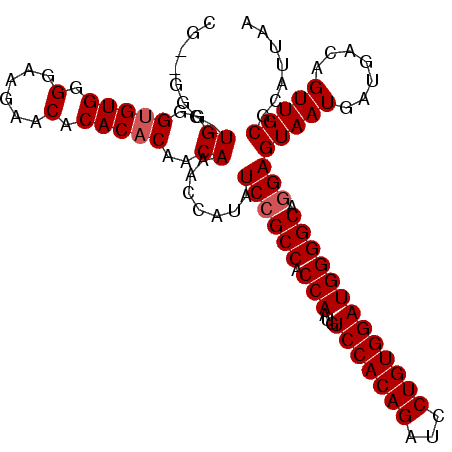

| Location | 12,344,032 – 12,344,129 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 75.90 |
| Mean single sequence MFE | -26.87 |
| Consensus MFE | -18.83 |
| Energy contribution | -19.50 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951229 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12344032 97 + 27905053 UCCACAGGAUCUGUGGACAUUGGUGGCGGAUAUAGUUUGUUGUGUGUGUUCUUCCUCACACACCA-CCC----AAUAAAA-ACACCCACCCACACACACGCAC--------- (((((((...)))))))....(((((..(.........((.(((((((........))))))).)-)..----.......-.)..))))).............--------- ( -27.01) >DroSec_CAF1 12779 108 + 1 UCCACAGGAUCUGUGGACAUUGGUGGCGGAUAUGGUUUGUUGUGUGUGUUCUUCCCCACACCCCA-CCCACCGAAUAAAA-AUACCCACCCACA--CACACACGCACGCACA (((((((...))))))).....((((((((((.....))))((((((((................-..............-..........)))--))))).))).)))... ( -25.30) >DroEre_CAF1 12743 107 + 1 UCCACAGGAUCUGUGGACAUUGGCGGCGGAUAUGGUUUGUUGUGUGUGUUU-UCCGCACCCCCCAAUA---AAAACGCC-CCCACCCACCCAUCCACCCACACACACGCACA ......((((..((((....(((.((((.......(((((((.(((((...-..)))))....)))))---))..))))-.))).))))..))))................. ( -28.30) >consensus UCCACAGGAUCUGUGGACAUUGGUGGCGGAUAUGGUUUGUUGUGUGUGUUCUUCCCCACACCCCA_CCC____AAUAAAA_ACACCCACCCACACACACACAC_CACGCACA (((((((...)))))))....(((((.(((((.....))))..(((((........))))).......................))))))...................... (-18.83 = -19.50 + 0.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:43 2006