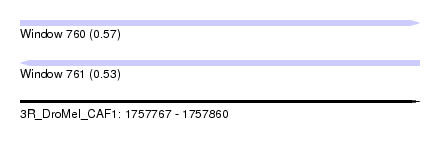
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,757,767 – 1,757,860 |
| Length | 93 |
| Max. P | 0.567696 |

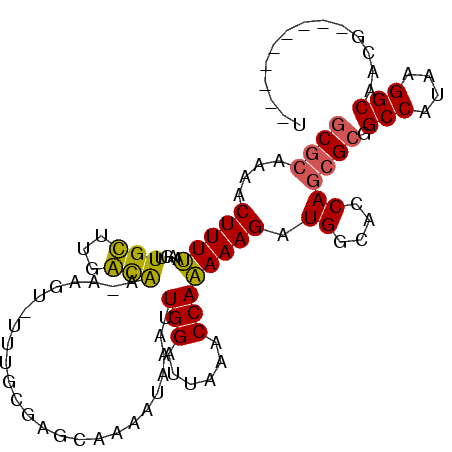
| Location | 1,757,767 – 1,757,860 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 79.17 |
| Mean single sequence MFE | -25.24 |
| Consensus MFE | -12.49 |
| Energy contribution | -12.80 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1757767 93 + 27905053 ----------CUUGGCCCUUUGGCCCCGCUGGUGCCAUCUUUCUGGAUUAAUCCAAUUUAUGUUGCUUGCCAA-ACUU--UAUGGAACAACUUGAAAGUUUUGCGC ----------...((((....))))......((((...(((((((((....))))......((((....(((.-....--..)))..))))..)))))....)))) ( -21.40) >DroVir_CAF1 79002 93 + 1 A---------CGCUGCCUUAUGGCCGCGCUGGUGCCAUCUUUUUGGUUUAAUCCAAUUUAUUUUGCUCACA--AACU--UUACCAAGCAACUUAAAAGUUUUGCGC .---------.((.(((....))).))(((((.((((......)))).....))).((((..(((((....--....--......)))))..))))......)).. ( -20.34) >DroPse_CAF1 57862 104 + 1 UUGCUGUUGGCCUGGCCUUAUGGCCGCGCUGGUGCCAUCUUUUUGGUUUAAUCCAAUUUAUGCUACUCGCAAA-ACUU-UUGUAAAACAACUUAAAAGUUUUGCGC ..((....(((.(((((....))))).)))((.((((......)))).....)).......))....((((((-((((-(((..........))))))))))))). ( -32.80) >DroGri_CAF1 68257 97 + 1 A---------CGUUGCCUUAUGGCCGCUCUGGUGCCAUCUUUUUGGUUUAAUCCAAUUUAUUUUGCUCAGAAAACUUUUGCGCCAAGCAACUUAAAAGUUUUGCGC .---------.((.(((....))).))(((((.((.......((((......))))........)))))))........((((.((((.........)))).)))) ( -22.36) >DroMoj_CAF1 72254 97 + 1 A---------CGCUGCCUUAUGGCCGCGCUGGUGCCAUCUUUUUGGUUUAAUCCAAUUUAUUUUCCGCACAGAAACUUUUUGCCGAGCAACUUAAAAGUUUGCCGC .---------(((.(((....))).)))((((((........((((......)))).........))).)))(((((((((((...)))....))))))))..... ( -21.73) >DroPer_CAF1 64356 104 + 1 UUGCUGUUGGCCUGGCCUUAUGGCCGCGCUGGUGCCAUCUUUUUGGUUUAAUCCAAUUUAUGCUACUCGCAAA-ACUU-UUGUAAAACAACUUAAAAGUUUUGCGC ..((....(((.(((((....))))).)))((.((((......)))).....)).......))....((((((-((((-(((..........))))))))))))). ( -32.80) >consensus A_________CGUGGCCUUAUGGCCGCGCUGGUGCCAUCUUUUUGGUUUAAUCCAAUUUAUGUUGCUCACAAA_ACUU_UUGCCAAACAACUUAAAAGUUUUGCGC ..............(((....))).(((((((.((((......)))).....)))............................(((((.........))))))))) (-12.49 = -12.80 + 0.31)

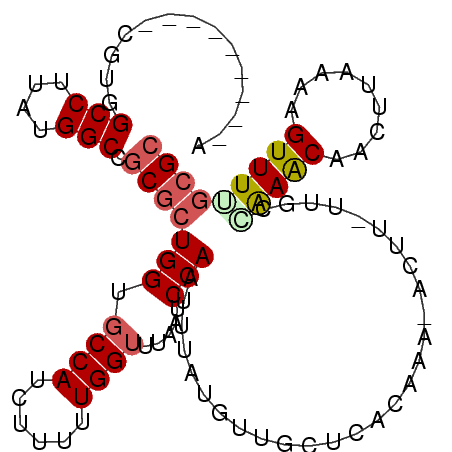
| Location | 1,757,767 – 1,757,860 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 79.17 |
| Mean single sequence MFE | -25.43 |
| Consensus MFE | -12.85 |
| Energy contribution | -12.22 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.534099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1757767 93 - 27905053 GCGCAAAACUUUCAAGUUGUUCCAUA--AAGU-UUGGCAAGCAACAUAAAUUGGAUUAAUCCAGAAAGAUGGCACCAGCGGGGCCAAAGGGCCAAG---------- .(((....(((((..(((((((((..--....-.)))..))))))......((((....))))))))).((....))))).((((....))))...---------- ( -27.80) >DroVir_CAF1 79002 93 - 1 GCGCAAAACUUUUAAGUUGCUUGGUAA--AGUU--UGUGAGCAAAAUAAAUUGGAUUAAACCAAAAAGAUGGCACCAGCGCGGCCAUAAGGCAGCG---------U ((.((.....((((..((((((.....--....--...))))))..))))((((......)))).....))))....((((.(((....))).)))---------) ( -21.30) >DroPse_CAF1 57862 104 - 1 GCGCAAAACUUUUAAGUUGUUUUACAA-AAGU-UUUGCGAGUAGCAUAAAUUGGAUUAAACCAAAAAGAUGGCACCAGCGCGGCCAUAAGGCCAGGCCAACAGCAA .((((((((((((..((......))))-))))-))))))....((.....((((......))))...(.((((........((((....))))..)))).).)).. ( -29.50) >DroGri_CAF1 68257 97 - 1 GCGCAAAACUUUUAAGUUGCUUGGCGCAAAAGUUUUCUGAGCAAAAUAAAUUGGAUUAAACCAAAAAGAUGGCACCAGAGCGGCCAUAAGGCAACG---------U .(((.((((((((..(((....)))..))))))))((((.((........((((......)))).......))..)))))))(((....)))....---------. ( -21.96) >DroMoj_CAF1 72254 97 - 1 GCGGCAAACUUUUAAGUUGCUCGGCAAAAAGUUUCUGUGCGGAAAAUAAAUUGGAUUAAACCAAAAAGAUGGCACCAGCGCGGCCAUAAGGCAGCG---------U .((((((.((....)))))).))((......(((((....))))).....((((......)))).......))....((((.(((....))).)))---------) ( -22.50) >DroPer_CAF1 64356 104 - 1 GCGCAAAACUUUUAAGUUGUUUUACAA-AAGU-UUUGCGAGUAGCAUAAAUUGGAUUAAACCAAAAAGAUGGCACCAGCGCGGCCAUAAGGCCAGGCCAACAGCAA .((((((((((((..((......))))-))))-))))))....((.....((((......))))...(.((((........((((....))))..)))).).)).. ( -29.50) >consensus GCGCAAAACUUUUAAGUUGCUUGACAA_AAGU_UUUGCGAGCAAAAUAAAUUGGAUUAAACCAAAAAGAUGGCACCAGCGCGGCCAUAAGGCAACG_________U ((((....(((((....(((...))).........................(((......)))))))).((....)))))).(((....))).............. (-12.85 = -12.22 + -0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:58 2006