
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,336,341 – 12,336,489 |
| Length | 148 |
| Max. P | 0.999666 |

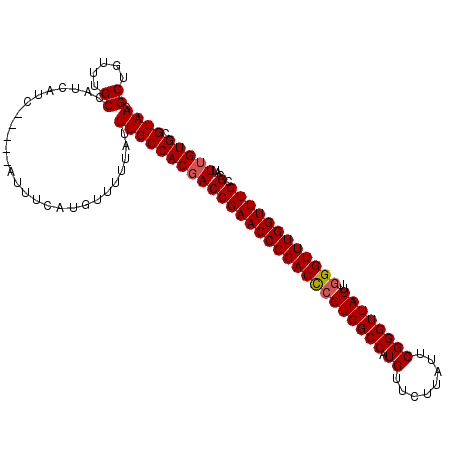
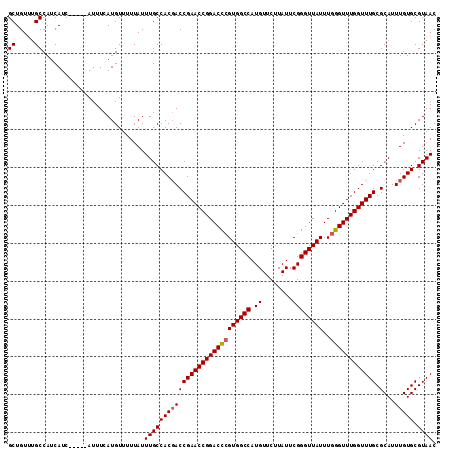
| Location | 12,336,341 – 12,336,449 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 95.43 |
| Mean single sequence MFE | -28.78 |
| Consensus MFE | -27.42 |
| Energy contribution | -27.66 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.03 |
| Structure conservation index | 0.95 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12336341 108 + 27905053 GCUGUUUGCCAUCAUC-----AUUUCAUGUUUUUAUUUGCCACGACCGAACCGGACCAGUGGCCAUGUUCUUAUUCGGGUUAUUUGGGUUUGGUUUGCGCAUUUGUGCGUAAC ((.....)).......-----...............(((((((((((((((((((((.((((((.((........))))))))...))))))))))).)...))))).)))). ( -27.30) >DroSec_CAF1 5179 112 + 1 GCUGU-UGCCAUCAUCGUUUCAUUUCAUGUUUUUAUUUGCCACGACCGAACCGGACCCGUGGCCAUGUUCUUAUUCGGGUUAUUUGGGUUUGGUUUGCGCAUUUGUGCGUAAC ...((-(((.....((((..((...............))..))))..(((((((((((((((((.((........))))))))..)))))))))))((((....))))))))) ( -30.76) >DroSim_CAF1 4421 113 + 1 GCUGUUUGCCAUCAUCAUUUCAUUUCAUGUUUUUAUUUGCCACGACCGAACCGGACCCGUGGCCAUGUUCUUAUUCGGGUUAUUUGGGUUUGGUUUGCGCAUUUGUGCGUAAC ((.....))...........................((((((((((((((((((((((((((((.((........))))))))..)))))))))))).)...))))).)))). ( -29.60) >DroEre_CAF1 4564 108 + 1 GCUGUUUGCCAUCAUC-----AUUUCAUGUUUUUAUUUGCCACGACCGAACCGGACCCGUGGCCAUGUUCUUAUUCGGGUUAUUUGGGUUUGGUUUGCGCAUUUGUGCGUAAC ((.....)).......-----...............((((((((((((((((((((((((((((.((........))))))))..)))))))))))).)...))))).)))). ( -29.60) >DroYak_CAF1 645 108 + 1 GCUGUUGGCCAUCAUC-----AUUUCAUGUUUUUAUUUGCCACUACCGAACCGGACUCGUGGCCAUGUUCUUAUUCGGGUUAUUUGGGUUUGGUUUGCGCAUUUGUGCGUAAC .....((((.......-----.................))))..((((((((.((...((((((.((........)))))))))).))))))))(((((((....))))))). ( -26.66) >consensus GCUGUUUGCCAUCAUC_____AUUUCAUGUUUUUAUUUGCCACGACCGAACCGGACCCGUGGCCAUGUUCUUAUUCGGGUUAUUUGGGUUUGGUUUGCGCAUUUGUGCGUAAC ((.....))...........................((((((((((((((((((((((((((((.((........))))))))..)))))))))))).)...))))).)))). (-27.42 = -27.66 + 0.24)

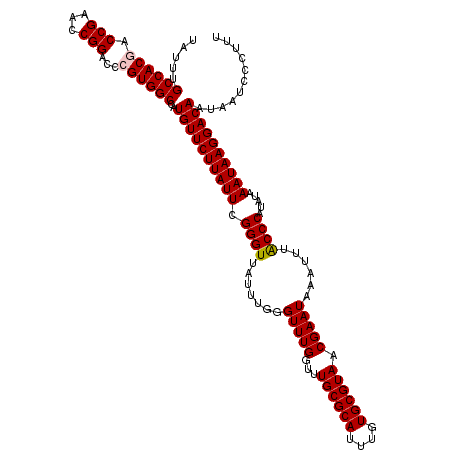
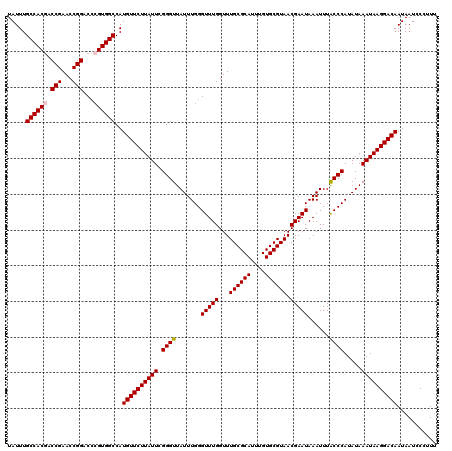
| Location | 12,336,369 – 12,336,489 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.50 |
| Mean single sequence MFE | -35.08 |
| Consensus MFE | -34.68 |
| Energy contribution | -34.84 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.99 |
| SVM decision value | 3.86 |
| SVM RNA-class probability | 0.999666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12336369 120 + 27905053 UAUUUGCCACGACCGAACCGGACCAGUGGCCAUGUUCUUAUUCGGGUUAUUUGGGUUUGGUUUGCGCAUUUGUGCGUAACGAAUAAAUUUACCCAUAUAAAUAAGGACAAUAAUCCCUUU .....(((((..(((...)))....)))))..((((((((((.((((.......(((((...((((((....)))))).)))))......)))).....))))))))))........... ( -34.22) >DroSec_CAF1 5211 120 + 1 UAUUUGCCACGACCGAACCGGACCCGUGGCCAUGUUCUUAUUCGGGUUAUUUGGGUUUGGUUUGCGCAUUUGUGCGUAACGAAUAAAUUUGCCCAUAUAAAUAAGGACAAUAAUCCCUUU .....((((((.(((...)))...))))))..((((((((((.((((.......(((((...((((((....)))))).)))))......)))).....))))))))))........... ( -36.12) >DroSim_CAF1 4454 120 + 1 UAUUUGCCACGACCGAACCGGACCCGUGGCCAUGUUCUUAUUCGGGUUAUUUGGGUUUGGUUUGCGCAUUUGUGCGUAACGAAUAAAUUUGCCCAUAUAAAUAAGGACAAUAAUCCCUUU .....((((((.(((...)))...))))))..((((((((((.((((.......(((((...((((((....)))))).)))))......)))).....))))))))))........... ( -36.12) >DroEre_CAF1 4592 120 + 1 UAUUUGCCACGACCGAACCGGACCCGUGGCCAUGUUCUUAUUCGGGUUAUUUGGGUUUGGUUUGCGCAUUUGUGCGUAACGAAUAAAUUUACCCAUAUAAAUAAGGACAAUAAUCCCUUU .....((((((.(((...)))...))))))..((((((((((.((((.......(((((...((((((....)))))).)))))......)))).....))))))))))........... ( -35.82) >DroYak_CAF1 673 120 + 1 UAUUUGCCACUACCGAACCGGACUCGUGGCCAUGUUCUUAUUCGGGUUAUUUGGGUUUGGUUUGCGCAUUUGUGCGUAACGAAUAAAUUUACCCAUAUAAAUAAGGACAAUAAUCCCUUU .....(((((..(((...)))....)))))..((((((((((.((((.......(((((...((((((....)))))).)))))......)))).....))))))))))........... ( -33.12) >consensus UAUUUGCCACGACCGAACCGGACCCGUGGCCAUGUUCUUAUUCGGGUUAUUUGGGUUUGGUUUGCGCAUUUGUGCGUAACGAAUAAAUUUACCCAUAUAAAUAAGGACAAUAAUCCCUUU .....((((((.(((...)))...))))))..((((((((((.((((.......(((((...((((((....)))))).)))))......)))).....))))))))))........... (-34.68 = -34.84 + 0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:23 2006