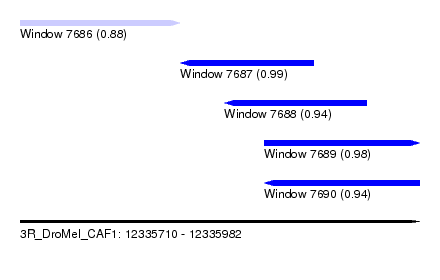
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,335,710 – 12,335,982 |
| Length | 272 |
| Max. P | 0.988802 |

| Location | 12,335,710 – 12,335,819 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 92.47 |
| Mean single sequence MFE | -26.17 |
| Consensus MFE | -23.81 |
| Energy contribution | -24.37 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875965 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12335710 109 + 27905053 GCAAAAGGGGUUGGGGGUUACGGGUCCUGGCUGCCAAAAAAA---AAA----AAAGUGAACAAAAACUAGGAAAGGGAAACGAGGGAACUUAAACACUUCUUGCAUUUUUCUUAUG ((((..(((((..(((.(....).)))..))).)).......---...----.(((((.................(....)(((....)))...))))).))))............ ( -23.00) >DroSec_CAF1 4525 113 + 1 GCCAAAGGGGUUGGGGGUUACGGGUCCUGGCUGCCAAAAAAA---AAAAAAAGAAGUGAAGAAAAACUGGGAAAGGGAAACGAGGGAACUUAAACACUUCUUGCAUUUUUCUUAUG ((....(((((..(((.(....).)))..))).)).......---.....((((((((.................(....)(((....)))...))))))))))............ ( -26.40) >DroSim_CAF1 3760 116 + 1 GCCAAAGGGGUUGGGGGUUACGGGUCCUGGCUGCCAAAAAAAAAAAAAAAAAGAAGUGAAGAAAAACUGGGAAAGGGAAACGAGGGAACUUAAACACUUCUUGCAUUUUUCUUAUG ((....(((((..(((.(....).)))..))).))...............((((((((.................(....)(((....)))...))))))))))............ ( -26.40) >DroEre_CAF1 3817 113 + 1 AC--AAGGGGUUGGGGGUUACGGGUCCUGGCUGCCAAAAAAAA-AAAAAAAAGAAGUGAAGAAAAGCUGGGAAAGGGAAACGAGGCAACUUAAACACUUCUUGCAUUUUCCUUAUG ..--((((((((((.(((((.(....)))))).))))......-......((((((((.................(....)(((....)))...)))))))).....))))))... ( -28.90) >consensus GCCAAAGGGGUUGGGGGUUACGGGUCCUGGCUGCCAAAAAAA___AAAAAAAGAAGUGAAGAAAAACUGGGAAAGGGAAACGAGGGAACUUAAACACUUCUUGCAUUUUUCUUAUG ....((((((((((.(((((.(....)))))).)))).............((((((((.................(....)(((....)))...)))))))).....))))))... (-23.81 = -24.37 + 0.56)



| Location | 12,335,819 – 12,335,910 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 91.22 |
| Mean single sequence MFE | -29.30 |
| Consensus MFE | -26.49 |
| Energy contribution | -27.30 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12335819 91 - 27905053 AACAACAA----GGCGACAAGAAGCCCAACAACAGCAGGGCAUCCCUGACAGCUUCGUCUGGAUUUUUGGGCGAGGCAAUAAAUAGUUGUCGUGC ........----.(((((((((.((((..........)))).)).......((((((((..(....)..)))))))).........))))))).. ( -31.30) >DroSec_CAF1 4638 85 - 1 AACAACAA----GGCGACAAGAAGCCCAACAACA------CAUCUCUGACAGCUUCGUCUGGAUUUUUGGGCGAGGCAAUAAAUAGUUGUCGUGC .(((((..----(((........)))........------...........((((((((..(....)..))))))))........)))))..... ( -23.90) >DroSim_CAF1 3876 91 - 1 AACAACAA----GGCGACAAGAAGCCCAACAACAACAGGGCAUCCCUGACAGCUUCGUCUGGAUUUUUGGGCGAGGCAAUAAAUAGUUGUCGUGC ........----.(((((((((.((((..........)))).)).......((((((((..(....)..)))))))).........))))))).. ( -31.30) >DroEre_CAF1 3930 95 - 1 AGCAACAACAAUGGCGACAAGGAGCCCAACAACAAGAGGGAAUCCCUGGCAGCUUCGUCUGGAUUUUUGGGCGAGGCAAUAAAUAGUUGUCGUGC ..........((((((((.....(((..........((((...))))))).((((((((..(....)..))))))))........)))))))).. ( -30.70) >consensus AACAACAA____GGCGACAAGAAGCCCAACAACAACAGGGCAUCCCUGACAGCUUCGUCUGGAUUUUUGGGCGAGGCAAUAAAUAGUUGUCGUGC .............(((((((((.((((..........)))).)).......((((((((..(....)..)))))))).........))))))).. (-26.49 = -27.30 + 0.81)


| Location | 12,335,849 – 12,335,946 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 91.25 |
| Mean single sequence MFE | -24.10 |
| Consensus MFE | -21.46 |
| Energy contribution | -21.90 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943918 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12335849 97 - 27905053 UUUCUUGAUGCCACUACAGAAGUAAGCAACAGCAACAACAACAA----GGCGACAAGAAGCCCAACAACAGCAGGGCAUCCCUGACAGCUUCGUCUGGAUU .((((((.((((..(((....))).((....))...........----)))).))))))((((..........))))((((..(((......))).)))). ( -27.20) >DroSec_CAF1 4668 91 - 1 UUUCUUGAUGCCACUACAGAAGUAAGCAACAGCAACAACAACAA----GGCGACAAGAAGCCCAACAACA------CAUCUCUGACAGCUUCGUCUGGAUU (((((((.((((..(((....))).((....))...........----)))).)))))))..........------.((((..(((......))).)))). ( -18.40) >DroSim_CAF1 3906 97 - 1 UUUCUUGAUGCCACUACAGAAGUAAGCAACAGCAACAACAACAA----GGCGACAAGAAGCCCAACAACAACAGGGCAUCCCUGACAGCUUCGUCUGGAUU .((((((.((((..(((....))).((....))...........----)))).))))))((((..........))))((((..(((......))).)))). ( -27.20) >DroEre_CAF1 3960 101 - 1 UUUCUUGAUGCCACUACAGAAGUAAGCAGCAGCAACAGCAACAACAAUGGCGACAAGGAGCCCAACAACAAGAGGGAAUCCCUGGCAGCUUCGUCUGGAUU (((((((.(((((.(((....))).((....))..............))))).)))))))(((..........)))(((((..(((......))).))))) ( -23.60) >consensus UUUCUUGAUGCCACUACAGAAGUAAGCAACAGCAACAACAACAA____GGCGACAAGAAGCCCAACAACAACAGGGCAUCCCUGACAGCUUCGUCUGGAUU .((((((.((((..(((....))).((....))...............)))).))))))((((..........))))((((..(((......))).)))). (-21.46 = -21.90 + 0.44)



| Location | 12,335,876 – 12,335,982 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 94.29 |
| Mean single sequence MFE | -33.65 |
| Consensus MFE | -32.01 |
| Energy contribution | -32.08 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979255 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12335876 106 + 27905053 CUGCUGUUGUUGGGCUUCUUGUCGCC----UUGUUGUUGUUGCUGUUGCUUACUUCUGUAGUGGCAUCAAGAAAGUUCUUUGAUAUUGCCGGCGGCAGGAACAACUUACA .....((.((((...(((((((((((----...........((..((((........))))..))(((((((....)).)))))......)))))))))))))))..)). ( -32.60) >DroSec_CAF1 4693 102 + 1 ----UGUUGUUGGGCUUCUUGUCGCC----UUGUUGUUGUUGCUGUUGCUUACUUCUGUAGUGGCAUCAAGAAAGUUCUUUGAUAUUGCCGGCGGCAGGAACAACUUACA ----.((.((((...(((((((((((----...........((..((((........))))..))(((((((....)).)))))......)))))))))))))))..)). ( -32.60) >DroSim_CAF1 3933 106 + 1 CUGUUGUUGUUGGGCUUCUUGUCGCC----UUGUUGUUGUUGCUGUUGCUUACUUCUGUAGUGGCAUCAAGAAAGUUCUUUGAUAUUGCCGGCGGCAGGAACAACUUACA ....(((.((((...(((((((((((----...........((..((((........))))..))(((((((....)).)))))......)))))))))))))))..))) ( -32.70) >DroEre_CAF1 3987 110 + 1 CUCUUGUUGUUGGGCUCCUUGUCGCCAUUGUUGUUGCUGUUGCUGCUGCUUACUUCUGUAGUGGCAUCAAGAAAGUUCUUUGAUAUUGCCGGCGGCAGGAACAACUUACA ............(((........)))...((((((.(((((((((((((........)))))((((((((((....)).))))...)))))))))))).))))))..... ( -36.70) >consensus CUGUUGUUGUUGGGCUUCUUGUCGCC____UUGUUGUUGUUGCUGUUGCUUACUUCUGUAGUGGCAUCAAGAAAGUUCUUUGAUAUUGCCGGCGGCAGGAACAACUUACA ....(((.((((....((((((((((...............((..((((........))))..))(((((((....)).)))))......)))))))))).))))..))) (-32.01 = -32.08 + 0.06)


| Location | 12,335,876 – 12,335,982 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 94.29 |
| Mean single sequence MFE | -24.75 |
| Consensus MFE | -23.46 |
| Energy contribution | -23.28 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12335876 106 - 27905053 UGUAAGUUGUUCCUGCCGCCGGCAAUAUCAAAGAACUUUCUUGAUGCCACUACAGAAGUAAGCAACAGCAACAACAACAA----GGCGACAAGAAGCCCAACAACAGCAG (((..((((((.(((..((.((((...((((.((....))))))))))..(((....))).))..))).)))))).....----(((........)))........))). ( -23.90) >DroSec_CAF1 4693 102 - 1 UGUAAGUUGUUCCUGCCGCCGGCAAUAUCAAAGAACUUUCUUGAUGCCACUACAGAAGUAAGCAACAGCAACAACAACAA----GGCGACAAGAAGCCCAACAACA---- (((..((((((.(((..((.((((...((((.((....))))))))))..(((....))).))..))).)))))).....----(((........)))..)))...---- ( -23.60) >DroSim_CAF1 3933 106 - 1 UGUAAGUUGUUCCUGCCGCCGGCAAUAUCAAAGAACUUUCUUGAUGCCACUACAGAAGUAAGCAACAGCAACAACAACAA----GGCGACAAGAAGCCCAACAACAACAG (((..((((((.(((..((.((((...((((.((....))))))))))..(((....))).))..))).)))))).....----(((........)))..)))....... ( -23.60) >DroEre_CAF1 3987 110 - 1 UGUAAGUUGUUCCUGCCGCCGGCAAUAUCAAAGAACUUUCUUGAUGCCACUACAGAAGUAAGCAGCAGCAACAGCAACAACAAUGGCGACAAGGAGCCCAACAACAAGAG (((..((((((.((((.((.((((...((((.((....))))))))))..(((....))).)).)))).)))))).........(((........)))..)))....... ( -27.90) >consensus UGUAAGUUGUUCCUGCCGCCGGCAAUAUCAAAGAACUUUCUUGAUGCCACUACAGAAGUAAGCAACAGCAACAACAACAA____GGCGACAAGAAGCCCAACAACAACAG (((..((((((.(((..((.((((...((((.((....))))))))))..(((....))).))..))).)))))).........(((........)))..)))....... (-23.46 = -23.28 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:21 2006