| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,754,926 – 1,755,040 |
| Length | 114 |
| Max. P | 0.686642 |

| Location | 1,754,926 – 1,755,040 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.06 |
| Mean single sequence MFE | -28.90 |
| Consensus MFE | -20.91 |
| Energy contribution | -20.52 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.686642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
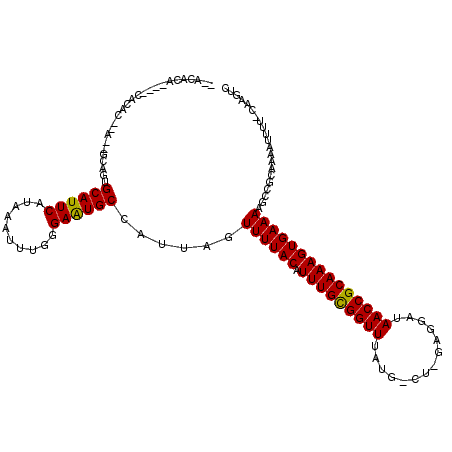
>3R_DroMel_CAF1 1754926 114 - 27905053 --GCACAAUUGCGCAC--AUUGCAUUGCAUUCAUAAAUUUCGGAAUGCCAUUAGUUUUACAUUUGCGGUUUGUG-CUCGGGGAUAACCGCAAAGUGAAAAGCCGCAAAAUUUC-CAAGUC --......((((((..--........((((((..........))))))......((((((.(((((((((.((.-(....).))))))))))))))))).).)))))......-...... ( -28.90) >DroVir_CAF1 76212 110 - 1 -CACACA----CACACUUG---UAGUGCAUUCAUAAAUUUGCGAGUGCCAUUAGUUUUACAUUUGCGGUUUAUG-CU-GAGGAUAACCGCAAAGUGAAAAGCCGCAAAAUUUUCCAAGUC -......----...(((((---((((((((((..........))))).)))))........((((((((((..(-((-..((....))....)))...))))))))))......))))). ( -26.30) >DroGri_CAF1 65316 102 - 1 UCAC--------------A---UAGUGCAUUCAUAAAUUUGUGAGUGCCAUUAGUUUUACAUUUGCGGUUUAUGGCU-GAGGAUAACCGCAAAGUGAAAAGCCGCAAAAUUUUCCAAGUC ....--------------.---(((((((((((((....)))))))).)))))........((((((((((.(.(((-..((....))....))).).))))))))))............ ( -29.50) >DroWil_CAF1 58693 109 - 1 -UACACA----CACAC--ACGACGUUGCAUUCAUAAAUUUUGGAAUGCCAUUAGUUUUACAUUUGUGGUUUGUG-CU-GAGGAUAACCGCAAAGUGAAAAGCCGCAA-AUUUU-CAAGUC -......----.....--..(((...((((((..........))))))............(((((((((((..(-((-..((....))....)))...)))))))))-))...-...))) ( -23.20) >DroMoj_CAF1 69417 113 - 1 CUACAUA----CAUAUGUGAAGCAGUGCAUUCAUAAAUUUGCGAGUGCCAUUAGUUUUACAUUUGCGGUUUAUG-CU-GAGGAUAACCGCAAAGUGAAAAGCCGCAAAAUUUU-CAAGUC .......----....(((((((((((((((((..........))))).)))).))))))))((((((((((..(-((-..((....))....)))...)))))))))).....-...... ( -30.30) >DroPer_CAF1 61697 109 - 1 --GAGCA----UACAC--GGGGCAUUGCAUUCAUAAAUUUUGGAAUGCCAUUAGUUUUACAUUUGCGGUUUGUG-CU-GGGGAUAACCGCAAAGUGAAAAGCCGCAAAAUGUU-CAAGUC --(((((----(...(--((((((((.((...........)).)))))).....((((((.(((((((((....-(.-...)..)))))))))))))))..)))....)))))-)..... ( -35.20) >consensus __ACACA____CACAC__A__GCAGUGCAUUCAUAAAUUUGGGAAUGCCAUUAGUUUUACAUUUGCGGUUUAUG_CU_GAGGAUAACCGCAAAGUGAAAAGCCGCAAAAUUUU_CAAGUC ..........................((((((..........))))))......((((((.(((((((((..............)))))))))))))))..................... (-20.91 = -20.52 + -0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:55 2006