
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,310,590 – 12,310,750 |
| Length | 160 |
| Max. P | 0.774698 |

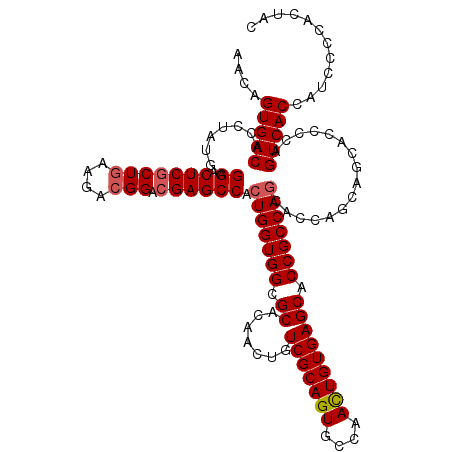
| Location | 12,310,590 – 12,310,710 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -54.90 |
| Consensus MFE | -52.22 |
| Energy contribution | -52.33 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.02 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.678927 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12310590 120 + 27905053 AGUGGCGCACGCAGGAGCGCUUCUCGAACCGCGUGUUCUGGUAGUGGGGAUGGUGACUGGGGUGCUGCUGGUGCUGGCGGUGCUCACAAUUGGCACUGCGACAGUUAUGCGCCACCAGUG .((((((((..(((.((((((((.....((((...........))))....(....).)))))))).)))..((((((((((((.......))))))))..))))..))))))))..... ( -52.50) >DroSec_CAF1 1883 120 + 1 AGUGGCGCACGCAGGAGCGCUUCUCGAACCGCGUGUUCUGGUAGUGGGGAUGGUGACUGGGGUGCUGCUGGUGUUGGCGGUGCUCACAGUUGGCACUGCGACAGUUGUGCGCCACCAGUG .(((((((((.(((.((((((((.....((((...........))))....(....).)))))))).))).(((((.(((((((.......))))))))))))...)))))))))..... ( -56.10) >DroSim_CAF1 1863 120 + 1 AGUGGCGCACGCAGGAGCGCUUCUCGAACCGCGUGUUCUGGUAGUGGGGAUGGUGACUGGGGUGCUGCUGGUGCUGGCGGUGCUCACAGUUGGCACUGCGACAGUUGUGCGCCACCAGUG .(((((((((.(((.((((((((.....((((...........))))....(....).)))))))).)))..((((((((((((.......))))))))..)))).)))))))))..... ( -56.10) >consensus AGUGGCGCACGCAGGAGCGCUUCUCGAACCGCGUGUUCUGGUAGUGGGGAUGGUGACUGGGGUGCUGCUGGUGCUGGCGGUGCUCACAGUUGGCACUGCGACAGUUGUGCGCCACCAGUG .(((((((((((((.((((((((....((((....((((......)))).))))....)))))))).)))..((((((((((((.......))))))))..))))))))))))))..... (-52.22 = -52.33 + 0.11)



| Location | 12,310,630 – 12,310,750 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -51.23 |
| Consensus MFE | -50.00 |
| Energy contribution | -50.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.774698 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12310630 120 + 27905053 GUAGUGGGGAUGGUGACUGGGGUGCUGCUGGUGCUGGCGGUGCUCACAAUUGGCACUGCGACAGUUAUGCGCCACCAGUGGCUCGUCCGUCUUCAGAGAGUCCUCAUAGGUGUCACUGUU .(((((((((((((.((((((((((.((((....(.((((((((.......)))))))).)))))...))))).))))).)).))))).......(((....))).......)))))).. ( -52.20) >DroSec_CAF1 1923 120 + 1 GUAGUGGGGAUGGUGACUGGGGUGCUGCUGGUGUUGGCGGUGCUCACAGUUGGCACUGCGACAGUUGUGCGCCACCAGUGGCUCGUCCGUCUUUAGCGAGUCCUCAUAGGUGUCACUGUU .(((((((((((((.((((((((((.((.(.(((((.(((((((.......)))))))))))).).))))))).))))).)).))))).............((.....))..)))))).. ( -51.00) >DroSim_CAF1 1903 120 + 1 GUAGUGGGGAUGGUGACUGGGGUGCUGCUGGUGCUGGCGGUGCUCACAGUUGGCACUGCGACAGUUGUGCGCCACCAGUGGCUCGUCCGUCUUCAGCGAGUCCUCAUAGGUGUCACUGUU .......(((((((.((((((((((.((((....(.((((((((.......)))))))).)))))...))))).))))).)).))))).....(((.((..((.....))..)).))).. ( -50.50) >consensus GUAGUGGGGAUGGUGACUGGGGUGCUGCUGGUGCUGGCGGUGCUCACAGUUGGCACUGCGACAGUUGUGCGCCACCAGUGGCUCGUCCGUCUUCAGCGAGUCCUCAUAGGUGUCACUGUU .(((((((((((((.((((((((((.((((....(.((((((((.......)))))))).)))))...))))).))))).)).))))).............((.....))..)))))).. (-50.00 = -50.00 + 0.00)



| Location | 12,310,630 – 12,310,750 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -37.70 |
| Consensus MFE | -34.83 |
| Energy contribution | -35.61 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.622995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12310630 120 - 27905053 AACAGUGACACCUAUGAGGACUCUCUGAAGACGGACGAGCCACUGGUGGCGCAUAACUGUCGCAGUGCCAAUUGUGAGCACCGCCAGCACCAGCAGCACCCCAGUCACCAUCCCCACUAC ....(((((........((.(((((((....)))).))))).(((((((.((.......(((((((....))))))))).)))))))................)))))............ ( -36.71) >DroSec_CAF1 1923 120 - 1 AACAGUGACACCUAUGAGGACUCGCUAAAGACGGACGAGCCACUGGUGGCGCACAACUGUCGCAGUGCCAACUGUGAGCACCGCCAACACCAGCAGCACCCCAGUCACCAUCCCCACUAC ....(((((........((.((((((......)).))))))..((((((.((.(....)(((((((....))))))))).)))))).................)))))............ ( -35.90) >DroSim_CAF1 1903 120 - 1 AACAGUGACACCUAUGAGGACUCGCUGAAGACGGACGAGCCACUGGUGGCGCACAACUGUCGCAGUGCCAACUGUGAGCACCGCCAGCACCAGCAGCACCCCAGUCACCAUCCCCACUAC ....(((((........((.(((((((....))).)))))).(((((((.((.(....)(((((((....))))))))).)))))))................)))))............ ( -40.50) >consensus AACAGUGACACCUAUGAGGACUCGCUGAAGACGGACGAGCCACUGGUGGCGCACAACUGUCGCAGUGCCAACUGUGAGCACCGCCAGCACCAGCAGCACCCCAGUCACCAUCCCCACUAC ....(((((........((.(((((((....))).)))))).(((((((.((.......(((((((....))))))))).)))))))................)))))............ (-34.83 = -35.61 + 0.78)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:51 2006