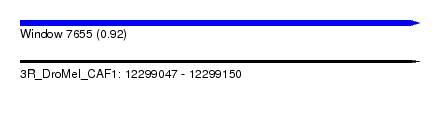
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,299,047 – 12,299,150 |
| Length | 103 |
| Max. P | 0.918295 |

| Location | 12,299,047 – 12,299,150 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 78.22 |
| Mean single sequence MFE | -27.03 |
| Consensus MFE | -20.57 |
| Energy contribution | -20.93 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918295 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12299047 103 + 27905053 CAUUUAAUCGUCCAUUGUUUUCCAUUUUUUCAGAAACAUAAACCAUGUAUGGAAUGCUGUAUGAGAGUGUGCAGCACUACGUGCAGGAGGAGUAUGGCGUGGA ........((.((((..((((((.((((....)))).........((((((...((((((((......))))))))...))))))))))))..)))))).... ( -28.10) >DroSec_CAF1 3048 87 + 1 CAUUUGA----------------AUUUUUUCAGAAACUUCAACCAUGUAUGGAAUGCUGUACGAGAGUGUGCAGCACUACGUGCAGGAGGAGUACGGCGUGGA ..(((((----------------(....))))))........((((((......(((((((((....)))))))))...(((((.......))))))))))). ( -29.00) >DroSim_CAF1 3093 87 + 1 CAUUUGA----------------AUUUUUUCAGAAACUUCAACCAUGUAUGGAAUGCUGUACGAGAGUGUGCAGCACUACGUGCAGGAGGAGUACGGCGUGGA ..(((((----------------(....))))))........((((((......(((((((((....)))))))))...(((((.......))))))))))). ( -29.00) >DroEre_CAF1 3235 102 + 1 UAUUAAUUAAUCCAUUCAUUUAAAUCUUUUCAGAAACACCAA-CAUGUAUGGAAUGCUGUACGAGAGUGUGCAGCACUACGUGCAGGAGGAGUACGGCGUGGA ..........(((((.(.......(((....))).((.((..-(.((((((...(((((((((....)))))))))...)))))))..)).))...).))))) ( -27.10) >DroYak_CAF1 3122 102 + 1 CAUUUAUU-GUCUAUUCACUCAAUUUUUUUCAGAAACAUCAACCAUGUAUGGGAUGCUGUACGAGAGUGUGCAGCACUACGUGCAGGAGGAGUACGGCGUGGA ........-((((((((.(((...............((((..(((....)))))))(((((((..((((.....)))).))))))))))))))).)))..... ( -29.00) >DroPer_CAF1 3231 90 + 1 ---UUAC----------UUUCACAGAUUCACAGAUCCAUCAAUCAUGUAUGGCAUGCUCUAUGAGAGUGUCCAGCACUAUGUGCAGGAGGAGUACGGCACACA ---.(((----------(((((.(((....((...((((((....)).))))..)).))).))))))))(((.((((...)))).)))............... ( -20.00) >consensus CAUUUAA___________UU_A_AUUUUUUCAGAAACAUCAACCAUGUAUGGAAUGCUGUACGAGAGUGUGCAGCACUACGUGCAGGAGGAGUACGGCGUGGA ..........................................((((((......((((((((......))))))))...(((((.......))))))))))). (-20.57 = -20.93 + 0.36)

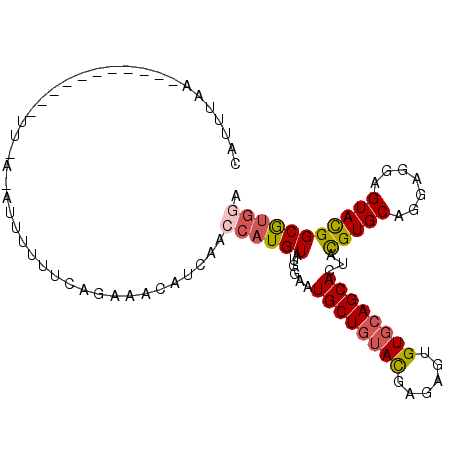
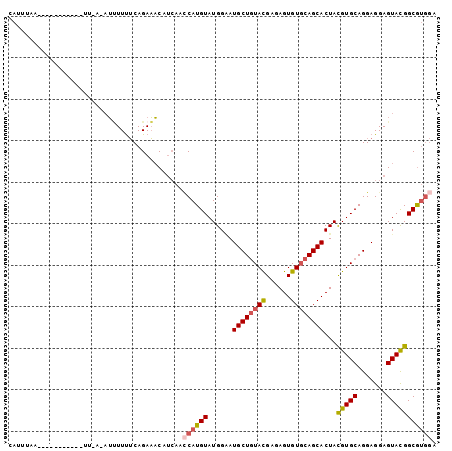
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:48 2006