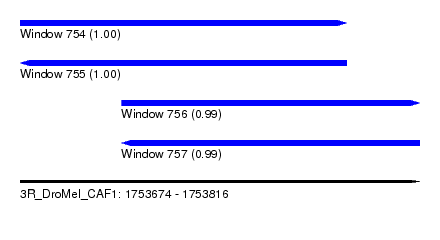
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,753,674 – 1,753,816 |
| Length | 142 |
| Max. P | 0.998576 |

| Location | 1,753,674 – 1,753,790 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 81.16 |
| Mean single sequence MFE | -37.24 |
| Consensus MFE | -35.01 |
| Energy contribution | -35.57 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.15 |
| SVM RNA-class probability | 0.998576 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1753674 116 + 27905053 GAUAGUUAAAAGAUCUUAAAAGAUUAAAAGAUAUCAGGCAGUCCAUCCGUCGCCAGGUGGCUCUCUUGUGCAAUUACUGUGGCGCACUCCGUAGAAGAGCCACCUGGCGUUAAAUG ((((.......(((((....)))))......))))........(((....(((((((((((((((((((((.((....)).)))))......)).))))))))))))))....))) ( -40.12) >DroSec_CAF1 63000 97 + 1 -----------GAUCUUA--------AAAGAUAUUAUGCAGUCCCUCCGUCGCCAGGUGGCUCUCUUGCGUAAUUACUGUGGCGCACCCAGUAGAAGACCCACCUGGCGUUAGGUA -----------.((((..--------..))))..............((..((((((((((.((((((((((.((....)).)))))......)).))).))))))))))...)).. ( -32.60) >DroSim_CAF1 65373 97 + 1 -----------GAUCUUA--------AAAGAUAUUAUGCAGUCCAUCCGUCGCCAGGUGGCUCUCUUGCGUAAUUACAGUGGCGCACCCAGUAGAAGAGCCACCUGGCGUUAGGUA -----------.((((..--------..))))..............((..(((((((((((((((((((((.((....)).)))))......)).))))))))))))))...)).. ( -39.00) >consensus ___________GAUCUUA________AAAGAUAUUAUGCAGUCCAUCCGUCGCCAGGUGGCUCUCUUGCGUAAUUACUGUGGCGCACCCAGUAGAAGAGCCACCUGGCGUUAGGUA ............(((((..........)))))..............((..(((((((((((((((((((((.((....)).)))))......)).))))))))))))))...)).. (-35.01 = -35.57 + 0.56)

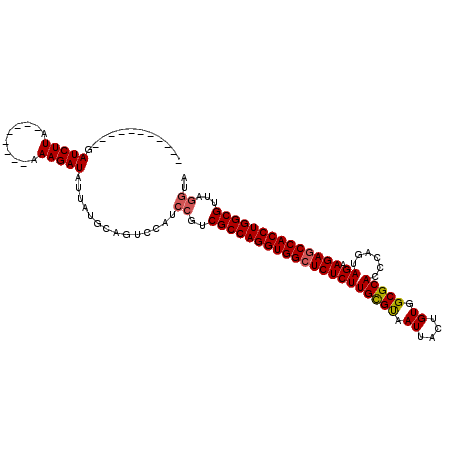
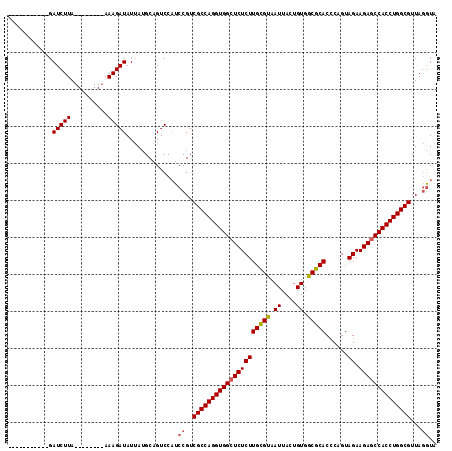
| Location | 1,753,674 – 1,753,790 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 81.16 |
| Mean single sequence MFE | -38.70 |
| Consensus MFE | -31.60 |
| Energy contribution | -31.93 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.61 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.94 |
| SVM RNA-class probability | 0.997817 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1753674 116 - 27905053 CAUUUAACGCCAGGUGGCUCUUCUACGGAGUGCGCCACAGUAAUUGCACAAGAGAGCCACCUGGCGACGGAUGGACUGCCUGAUAUCUUUUAAUCUUUUAAGAUCUUUUAACUAUC (((((..((((((((((((((((....))(((((..........)))))...))))))))))))))..)))))..............((((((....))))))............. ( -40.80) >DroSec_CAF1 63000 97 - 1 UACCUAACGCCAGGUGGGUCUUCUACUGGGUGCGCCACAGUAAUUACGCAAGAGAGCCACCUGGCGACGGAGGGACUGCAUAAUAUCUUU--------UAAGAUC----------- ..(((..((((((((((.(((..(((((((....)).)))))....(....)))).))))))))))....)))...........((((..--------..)))).----------- ( -38.50) >DroSim_CAF1 65373 97 - 1 UACCUAACGCCAGGUGGCUCUUCUACUGGGUGCGCCACUGUAAUUACGCAAGAGAGCCACCUGGCGACGGAUGGACUGCAUAAUAUCUUU--------UAAGAUC----------- ..((...((((((((((((((..(((..((....))...)))....(....)))))))))))))))..))..............((((..--------..)))).----------- ( -36.80) >consensus UACCUAACGCCAGGUGGCUCUUCUACUGGGUGCGCCACAGUAAUUACGCAAGAGAGCCACCUGGCGACGGAUGGACUGCAUAAUAUCUUU________UAAGAUC___________ .......(((((((((((((((........(((......))).........)))))))))))))))..................(((((..........)))))............ (-31.60 = -31.93 + 0.33)

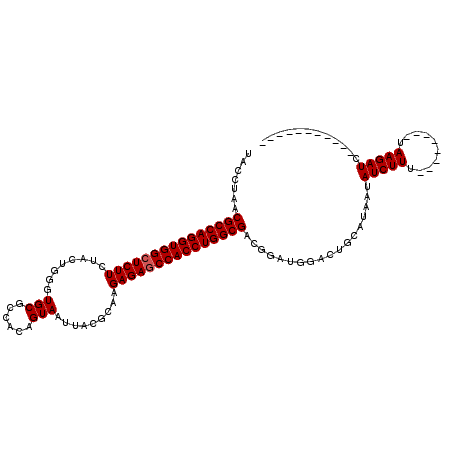
| Location | 1,753,710 – 1,753,816 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.78 |
| Mean single sequence MFE | -43.31 |
| Consensus MFE | -37.11 |
| Energy contribution | -38.33 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.40 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.994637 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1753710 106 + 27905053 GGCAGUCCAUCCGUCGCCAGGUGGCUCUCUUGUGCAAUUACUGUGGCGCACUCCGUAGAAGAGCCACCUGGCGUUAAAUG-ACAAACUGGA------UAUGUUCCAUGGCCUA------- (((.(((.......(((((((((((((((((((((.((....)).)))))......)).))))))))))))))......)-))....((((------.....))))..)))..------- ( -46.82) >DroSec_CAF1 63017 120 + 1 UGCAGUCCCUCCGUCGCCAGGUGGCUCUCUUGCGUAAUUACUGUGGCGCACCCAGUAGAAGACCCACCUGGCGUUAGGUAUACAAACUGUACAGUAAUCUGUUCCAUGAAUUAGUUUUAA .(((((....((..((((((((((.((((((((((.((....)).)))))......)).))).))))))))))...)).......)))))((((....)))).................. ( -38.30) >DroSim_CAF1 65390 120 + 1 UGCAGUCCAUCCGUCGCCAGGUGGCUCUCUUGCGUAAUUACAGUGGCGCACCCAGUAGAAGAGCCACCUGGCGUUAGGUAUACGAACUGUACAGUAAUCUGUUCCAUGAAUUAGUUUUAA .((((((...((..(((((((((((((((((((((.((....)).)))))......)).))))))))))))))...)).....).)))))((((....)))).................. ( -44.80) >consensus UGCAGUCCAUCCGUCGCCAGGUGGCUCUCUUGCGUAAUUACUGUGGCGCACCCAGUAGAAGAGCCACCUGGCGUUAGGUAUACAAACUGUACAGUAAUCUGUUCCAUGAAUUAGUUUUAA ((((((....((..(((((((((((((((((((((.((....)).)))))......)).))))))))))))))...)).......))))))............................. (-37.11 = -38.33 + 1.23)


| Location | 1,753,710 – 1,753,816 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.78 |
| Mean single sequence MFE | -46.33 |
| Consensus MFE | -36.36 |
| Energy contribution | -36.36 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.11 |
| Mean z-score | -4.15 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.994260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1753710 106 - 27905053 -------UAGGCCAUGGAACAUA------UCCAGUUUGU-CAUUUAACGCCAGGUGGCUCUUCUACGGAGUGCGCCACAGUAAUUGCACAAGAGAGCCACCUGGCGACGGAUGGACUGCC -------..(((...(((.....------)))((((..(-(......((((((((((((((((....))(((((..........)))))...))))))))))))))...))..))))))) ( -49.30) >DroSec_CAF1 63017 120 - 1 UUAAAACUAAUUCAUGGAACAGAUUACUGUACAGUUUGUAUACCUAACGCCAGGUGGGUCUUCUACUGGGUGCGCCACAGUAAUUACGCAAGAGAGCCACCUGGCGACGGAGGGACUGCA ..................((((....)))).((((((.(...((...((((((((((.(((..(((((((....)).)))))....(....)))).))))))))))..))).)))))).. ( -43.00) >DroSim_CAF1 65390 120 - 1 UUAAAACUAAUUCAUGGAACAGAUUACUGUACAGUUCGUAUACCUAACGCCAGGUGGCUCUUCUACUGGGUGCGCCACUGUAAUUACGCAAGAGAGCCACCUGGCGACGGAUGGACUGCA ..................((((....)))).((((((((...((...((((((((((((((..(((..((....))...)))....(....)))))))))))))))..)))))))))).. ( -46.70) >consensus UUAAAACUAAUUCAUGGAACAGAUUACUGUACAGUUUGUAUACCUAACGCCAGGUGGCUCUUCUACUGGGUGCGCCACAGUAAUUACGCAAGAGAGCCACCUGGCGACGGAUGGACUGCA ...............................((((((((...((...(((((((((((((((........(((......))).........)))))))))))))))..)))))))))).. (-36.36 = -36.36 + 0.01)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:54 2006