
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,261,786 – 12,261,898 |
| Length | 112 |
| Max. P | 0.705118 |

| Location | 12,261,786 – 12,261,898 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.82 |
| Mean single sequence MFE | -33.15 |
| Consensus MFE | -19.31 |
| Energy contribution | -19.23 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705118 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12261786 112 - 27905053 UGAGCACCAGGACUUCAAGGAGGAGAUGAAGCGUCUGCGCAAGCUGCGCGGCAAGGCGCCUCCCAAGAAGGGCGAGGGCAAGCGGGCCUCAAAGAAGUAG------UAAAAUAA--AUAU ...........(((((...((((.......((((..((....)).)))).((...((.((((((......)).))))))..))...))))...)))))..------........--.... ( -36.30) >DroVir_CAF1 2893 116 - 1 UGAGCAUCAGGACUUUAAGGAGGAAAUGAAACGCUUACGCAAGCUGCGUGGAAAAGCGCCGCCCAAGAAGGGCGAGGGCAAACGUGCAACAAAGAAAUAGAUC--AUUUACAAC--AUAC ...((.(((...(((....)))....))).(((.((((((.....))))))....((.((((((.....))))).).))...)))))................--.........--.... ( -27.00) >DroPse_CAF1 5073 115 - 1 CGAGCAUCAGGACUUCAAGGAGGAAAUGAAACGUUUGCGCAAGUUGCGUGGCAAGGCGGCUCCCAAGAAGGGAGAGGGCAAGCGCGCAUCCAAGAAGUAGAC---AUAAAUAAA--AUAC .........(.(((((..(((.....((...(((((((.(..(((((.(....).)))))((((.....))))..).)))))))..)))))..)))))...)---.........--.... ( -30.80) >DroGri_CAF1 3235 118 - 1 UGAGCAUCAGGACUUCAAGGAGGAAAUGAAACGCUUGCGCAAGCUGCGUGGCAAGGCGCCGCCCAAAAAGGGCGAGGGCAAACGUGCCUCCAAGAAAUAGAUGUAAUAAAAAAA--AUAU ...(((((...(.(((..(((((...((..((((..((....)).))))..))..((.((((((.....))))).).)).......)))))..))).).)))))..........--.... ( -41.80) >DroMoj_CAF1 3091 111 - 1 UGAGCACCAGGACUUCAGGGAGGAAAUGAAGCGUUUGCGAAAACUGCGCGGCAAAGCACCUCCUAAGAAGGGCGAGGGCAAACGAUCAACUAAGAAAUAGA---------CAACAGAUUC ...((.((.(..((((.((((((.......((((((....)))).))((......)).))))))..))))..)..))))......................---------.......... ( -26.30) >DroAna_CAF1 4925 112 - 1 UGAGCACCAGGAUUUCAAGGAGGAAAUGAAGCGGUUGCGCAAGCUGCGUGGCAAGGCGCCUCCCAAAAAGGGAGAGGGCAAACGGGCCUCUAAGAAAUAA------UGUACAAA--AUAU .......((..(((((..(((((...((.....((..(((.....)))..))...((.((((((.....))))).).))...))..)))))..)))))..------))......--.... ( -36.70) >consensus UGAGCACCAGGACUUCAAGGAGGAAAUGAAACGUUUGCGCAAGCUGCGUGGCAAGGCGCCUCCCAAGAAGGGCGAGGGCAAACGGGCAUCAAAGAAAUAGA_____UAAACAAA__AUAC ...((..((...(((....)))....))..((((..((....)).)))).))...((.((((((.....))))).).))......................................... (-19.31 = -19.23 + -0.08)
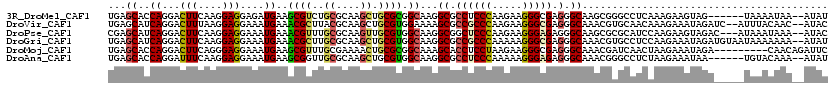
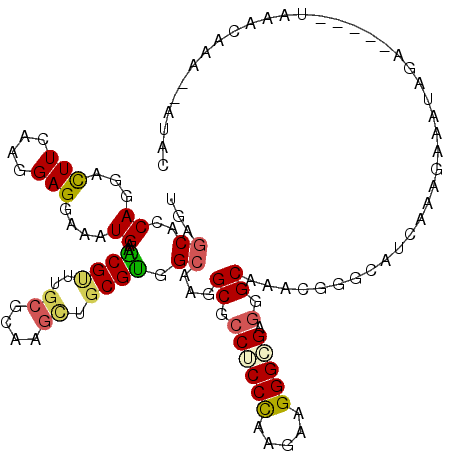

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:40 2006