
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,254,022 – 12,254,133 |
| Length | 111 |
| Max. P | 0.555101 |

| Location | 12,254,022 – 12,254,133 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.97 |
| Mean single sequence MFE | -24.70 |
| Consensus MFE | -16.75 |
| Energy contribution | -16.62 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.555101 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12254022 111 + 27905053 -----CGCGGAUUUUUCGCUAGCAUUUAUGCUUAAAUGCGCUGCAAUUGUUCUUAAAACGCUGCCAACCUGCACGCGAUGAUGGAAAACCUCAACGAUUUCCAUUUUAUU--CUGGCC-- -----.(((((...)))))..(((((((....)))))))(((................((((((......))).)))..((((((((.(......).)))))))).....--..))).-- ( -26.20) >DroVir_CAF1 84526 110 + 1 -----UCUGAACUUUUCGCCUGCAUUUAUGCUUAAAUGCGCUGCAAUUGUUCCUAAAAUGCAGCCA--CUGCACGCGAUGAUGGAAAACUUGAACGAUUUUCAUUUCUUUUU-UCUUU-- -----...(((....((((.((((.....((......))((((((.((.......)).))))))..--.)))).)))).((((((((.(......).))))))))......)-))...-- ( -25.10) >DroGri_CAF1 96577 111 + 1 -----UCUGAACUUUUCGCCUGCAUUUAUGCUUAAAUGCACUGCAAUUGUUUCUAAAAUGCAGCCA--CUGCACGCGAUGAUGGAAAACUUCAACGAUUUUCAUUUUUUU--CUUUUUUU -----...(((....((((.((((....(((......)))(((((.((.......)).)))))...--.)))).)))).((((((((.(......).))))))))...))--)....... ( -22.20) >DroMoj_CAF1 93073 113 + 1 -----UCUGAACUUUUCGCCUGCAUUUAUGCUUAAAUGCGCUGCAAUUGUUCCUAAAAUGCAGCCG--CGGCACGCGAUGAUGGAAAACUUGAACGAUUUUCAUUUAUUUUUCUCUUUGC -----............(((.(((((((....)))))))((((((.((.......)).))))))..--.)))..((((.((.((((((..((((.....))))....)))))))).)))) ( -29.20) >DroAna_CAF1 68116 116 + 1 AGUGAGGCAAACUUUUCGCUUGCAUUUAUGCUUAAAUGCACUGCAAUUGUUCUUAAAACGCUGCCAGGCUGCACGCGAUGAUGGAAAACUUCAACGAUUUUCAUUUUAUU--CUGGCC-- .....(((.........((.((((((((....))))))))..))....(((.....))))))(((((((.....))...((((((((.(......).)))))))).....--))))).-- ( -26.60) >DroPer_CAF1 76876 103 + 1 -----CGCGAACUUUUCGCCUGCAUUUAUGCUUAAAUGCCCAACAAUUGUUCUUAAAACGCU--------GCACGCGAUGAUGGAAAACUUCAACGAUUUUCAUUUUAUA--CUGGCC-- -----.((((.....))))..(((((((....)))))))(((................(((.--------....)))..((((((((.(......).)))))))).....--.)))..-- ( -18.90) >consensus _____UCCGAACUUUUCGCCUGCAUUUAUGCUUAAAUGCGCUGCAAUUGUUCCUAAAACGCAGCCA__CUGCACGCGAUGAUGGAAAACUUCAACGAUUUUCAUUUUAUU__CUGGCC__ ...............((((..(((((((....)))))))....................((.........))..)))).((((((((.(......).))))))))............... (-16.75 = -16.62 + -0.14)
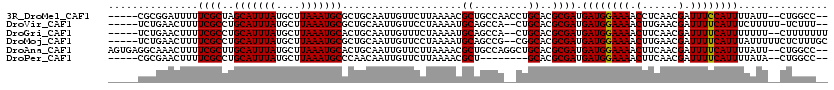
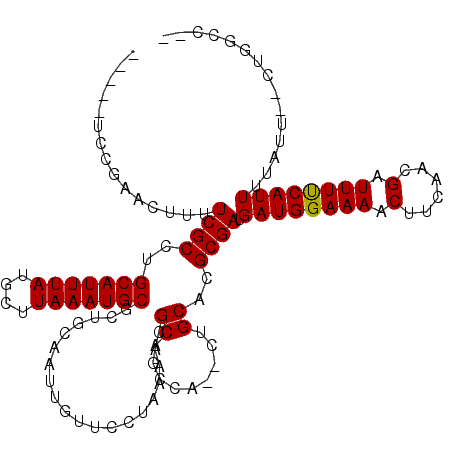

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:37 2006