| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,208,748 – 12,208,843 |
| Length | 95 |
| Max. P | 0.508998 |

| Location | 12,208,748 – 12,208,843 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 77.14 |
| Mean single sequence MFE | -28.88 |
| Consensus MFE | -16.13 |
| Energy contribution | -16.38 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.56 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.508998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
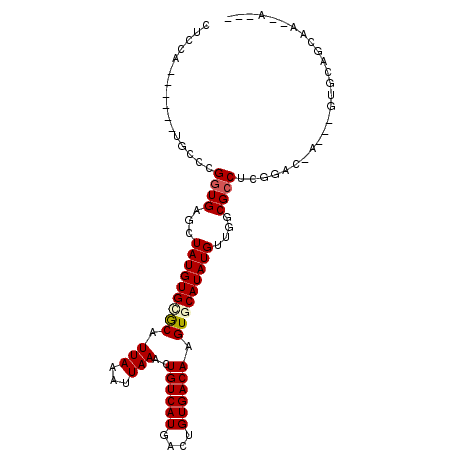
>3R_DroMel_CAF1 12208748 95 + 27905053 CUCCA-----AUGCCCGGUGAGCUAUGUGCGCAUUAAAUUAAACUGUCAUGACUGUGACAAGUGCAUAUGUUGGCGCCUCGGAC-A---GUGCAACAAACAAAU ...((-----.((.((((.(.(((((((((((.(((...)))..((((((....)))))).))))))))...))).).)))).)-)---.))............ ( -22.80) >DroVir_CAF1 29785 92 + 1 GUCUAGGUUU-GGCUGGGUGAGCUAUGUGUGCAUUAAAUUAAACUGUCAUGAGUGUGACAAGUGCAUAUGUUCGCGCCUGGC-C-ACAU--GCACC-A------ .....(((.(-(((..((((....(((((((((((.........((((((....)))))))))))))))))...))))..))-)-)...--..)))-.------ ( -34.60) >DroPse_CAF1 23309 95 + 1 CUCCA-----AUGCCCGGUGAGCUAUGUGGGCAUUAAAUUAAACUGUCAUGACUGUGACAAGUGCAUAUGUUGGCGCCUCGGAC-A---GUGCAGGAGAGAGAG (((((-----(((((((..........)))))))).........((((((....))))))..(((((.((((.(.....).)))-)---)))))))))...... ( -29.10) >DroWil_CAF1 25652 94 + 1 GCCCG------AGCUGGGUGAGCUAUGUGCACAUUAAAUUAAACUGUCAUGACUGUGACAAGUGCAUAUGUUGGCGCCUUGGACUA---CUACAACAA-CUACU ..(((------((..(.((.(((.((((((((.(((...)))..((((((....)))))).))))))))))).)).))))))....---.........-..... ( -25.60) >DroMoj_CAF1 32368 92 + 1 GCCUAGCUCU-GACCGGCUGAGCUAUGUGUGCAUUAAAUUAAACUGUCAUGAGUGUGACAAGUGCAUAUGUUCGCGCCUGGC-C-ACAA--GCGGC-A------ (((..(((.(-(.((((((((((...(((((((((.........((((((....)))))))))))))))))))).))).)).-)-)..)--)))))-.------ ( -32.10) >DroPer_CAF1 23156 93 + 1 CUCCA-----AUGCCCGGUGAGCUAUGUGGGCAUUAAAUUAAACUGUCAUGACUGUGACAAGUGCAUAUGUUGGCGCCUCGGAC-A---GUGCAGGAGAGAG-- (((((-----(((((((..........)))))))).........((((((....))))))..(((((.((((.(.....).)))-)---)))))))))....-- ( -29.10) >consensus CUCCA______UGCCCGGUGAGCUAUGUGCGCAUUAAAUUAAACUGUCAUGACUGUGACAAGUGCAUAUGUUGGCGCCUCGGAC_A___GUGCAGCAA__A___ ................((((...(((((((((.(((...)))..((((((....)))))).)))))))))....)))).......................... (-16.13 = -16.38 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:24 2006