| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,206,995 – 12,207,091 |
| Length | 96 |
| Max. P | 0.636147 |

| Location | 12,206,995 – 12,207,091 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 77.95 |
| Mean single sequence MFE | -24.08 |
| Consensus MFE | -13.00 |
| Energy contribution | -12.78 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.636147 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12206995 96 + 27905053 GUUGGCAUUAGCCGACAUAAAUGCUAAGUGGCGUCAUCAUUUAAAACGCAUCAAC--AGCCCG-C--AAGGAUGUGGUGCUG-CCAUGGCAUAACAUA--CAUA ((((((....))))))....(((((..((((((....).........((((((.(--(..((.-.--..)).)))))))).)-)))))))))......--.... ( -33.80) >DroVir_CAF1 26748 90 + 1 GUUGGUAUUAACCGACAUAAACGCUAAGUGGCGUCAUCAUUUAAAACGCAUCAACACAGCCCG-CCAAAGGAUAUGGAUAUGGGUUUGGAA------------- ((((((....))))))....(((((....)))))....................((.(((((.-(((.......)))....)))))))...------------- ( -20.20) >DroEre_CAF1 21460 91 + 1 GUUGGCAUUAGCCGACAUAAAUGCUAAGUGGCGUCAUCAUUUAAAACGCAUCAAC--AGCCCG-C--AAGGAUGUGGUGCUG-CCAUGGCAUA-CAUA------ ((((((....))))))....(((((..((((((....).........((((((.(--(..((.-.--..)).)))))))).)-))))))))).-....------ ( -33.80) >DroWil_CAF1 23220 94 + 1 GUUGGCAUUAGUCGACAUAAACGCUAAGUGGCGUCAUCAUUUAAAACGCAUCAAC--AGCCCG-C-AAAGGAUAUGGAUAUGGAUAUGG------AUAUGAAUG ((((((....)))))).....((((....))))((((.(((((...((.(((...--..((..-.-...)).....))).))....)))------))))))... ( -17.40) >DroAna_CAF1 21122 85 + 1 GUUGGCAUUAGCCGACAUAAAUGCUAAGUGGCGUCAUCAUUUAAAACGCAUCAAC--AGGCCGCC--AAGGAUAUGGUCCU-------------CAUA--CACA ((((((....))))))...........(((((((...........))))......--(((..(((--(......)))))))-------------....--))). ( -20.70) >DroPer_CAF1 21021 86 + 1 AUUGGCAUUAGCCGACAUAAAUGCUAAGUGGCGUCAUCAUUUAAAACGCAUCAACACAGCCUC-C-AAAGGAUAUGGCCCC------AGCAUA-C--------- .(((((....))))).....(((((..((.((((...........))))....))...(((((-(-...)))...)))...------))))).-.--------- ( -18.60) >consensus GUUGGCAUUAGCCGACAUAAAUGCUAAGUGGCGUCAUCAUUUAAAACGCAUCAAC__AGCCCG_C__AAGGAUAUGGUCCUG___AUGGCA___CAUA______ ((((((....))))))......(((..((.((((...........))))....))..)))((.......))................................. (-13.00 = -12.78 + -0.22)

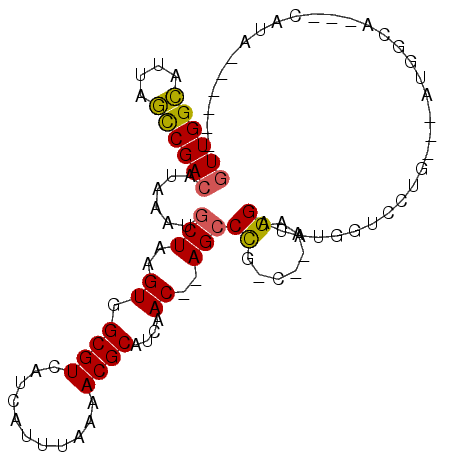

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:23 2006