
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,188,406 – 12,188,651 |
| Length | 245 |
| Max. P | 0.990382 |

| Location | 12,188,406 – 12,188,526 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.71 |
| Mean single sequence MFE | -33.22 |
| Consensus MFE | -31.93 |
| Energy contribution | -32.02 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.990382 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12188406 120 + 27905053 UUGAGAGAUCCAUGAUUUCAUAGAUUGUUCGGAAUGCUGGGAAAAUAUUUUUCACGGACUGAGCGGAAGCUUCUGCACAUGUACAUACAUAGGUGUAGAUCCAGUGGCAGAUUUCGAUAA ....((((((...)))))).........(((((((.((((((((....)))))....(((((((....)))(((((((((((....))))..)))))))..))))..))))))))))... ( -33.10) >DroSec_CAF1 2210 116 + 1 UUGAGAGAUCCAUGAUUUCAUAGAUUGUUCGGAAUGCUGGGAAAAUGUUUUUCACGGACUGGGCGGAAGCUUCUGCACACGUA----CAUAGGUGUAGAUCCAGUGGCAGAUUUCGAUAA ....((((((...)))))).........(((((((.((((((((....)))))....(((((((....)).(((((((.(...----....)))))))).)))))..))))))))))... ( -34.30) >DroSim_CAF1 2209 116 + 1 UUGAGAGAUCCAUGAUUUCAUAGAUUGUUCGGAAUGCUGGGAAAAUAUUUUUCACGGACUGGGCGGAAGCUUCUGCACAUGUA----CAUAGGUGUAGAUCCAGUGGCAGAUUUCGAUAA ....((((((...)))))).........(((((((.((((((((....)))))....(((((((....)).(((((((.....----.....))))))).)))))..))))))))))... ( -33.40) >DroEre_CAF1 2459 112 + 1 GUCAGAGAUUCAUGAUUUCAUAGAUUGUUCGGAAUGCUGGGAAAAUAUUUUUCACGGACUGGGCGGAAGCUUCUGCACAUG--------UACGUGUAGAUCCAGUGGCAGAUUUCGAUAA ......(((..(((....)))..)))..(((((((.((((((((....)))))....(((((((....)).(((((((...--------...))))))).)))))..))))))))))... ( -33.20) >DroYak_CAF1 2153 116 + 1 GUCAGAGAUUCAUGAUUUCAUAGAUUGUUCGGAAUGCUGGGAAAAUAUUUUUCACGGACUGGGCGGAAGCUUCUGCACAUGUA----UGUACGUGUAGAUCCAGUGGCAGAUUUCGAUAA ......(((..(((....)))..)))..(((((((.((((((((....)))))....(((((((....)).(((((((.....----.....))))))).)))))..))))))))))... ( -32.10) >consensus UUGAGAGAUCCAUGAUUUCAUAGAUUGUUCGGAAUGCUGGGAAAAUAUUUUUCACGGACUGGGCGGAAGCUUCUGCACAUGUA____CAUAGGUGUAGAUCCAGUGGCAGAUUUCGAUAA ....((((((...)))))).........(((((((.((((((((....)))))....(((((((....)).(((((((.(((......))).))))))).)))))..))))))))))... (-31.93 = -32.02 + 0.09)



| Location | 12,188,446 – 12,188,546 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 94.26 |
| Mean single sequence MFE | -26.34 |
| Consensus MFE | -24.89 |
| Energy contribution | -24.86 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896407 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12188446 100 + 27905053 GAAAAUAUUUUUCACGGACUGAGCGGAAGCUUCUGCACAUGUACAUACAUAGGUGUAGAUCCAGUGGCAGAUUUCGAUAAGUCAUUGGGCUAUAAUUUGC ............((((((...(((....)))(((((((((((....))))..)))))))))).)))(((((((......((((....))))..))))))) ( -26.70) >DroSec_CAF1 2250 96 + 1 GAAAAUGUUUUUCACGGACUGGGCGGAAGCUUCUGCACACGUA----CAUAGGUGUAGAUCCAGUGGCAGAUUUCGAUAAGUCAUUGGGCUAUAAUUUGC ((((.....))))....(((((((....)).(((((((.(...----....)))))))).))))).(((((((......((((....))))..))))))) ( -26.60) >DroSim_CAF1 2249 96 + 1 GAAAAUAUUUUUCACGGACUGGGCGGAAGCUUCUGCACAUGUA----CAUAGGUGUAGAUCCAGUGGCAGAUUUCGAUAAGUCAUUGGGCUAUAAUUUGC ((((.....))))....(((((((....)).(((((((.....----.....))))))).))))).(((((((......((((....))))..))))))) ( -25.70) >DroEre_CAF1 2499 92 + 1 GAAAAUAUUUUUCACGGACUGGGCGGAAGCUUCUGCACAUG--------UACGUGUAGAUCCAGUGGCAGAUUUCGAUAAGUCAUUGGGCUAUAAUUUGC ((((.....))))....(((((((....)).(((((((...--------...))))))).))))).(((((((......((((....))))..))))))) ( -26.90) >DroYak_CAF1 2193 96 + 1 GAAAAUAUUUUUCACGGACUGGGCGGAAGCUUCUGCACAUGUA----UGUACGUGUAGAUCCAGUGGCAGAUUUCGAUAAGUCAUUGGGCUAUAAUUUGC ((((.....))))....(((((((....)).(((((((.....----.....))))))).))))).(((((((......((((....))))..))))))) ( -25.80) >consensus GAAAAUAUUUUUCACGGACUGGGCGGAAGCUUCUGCACAUGUA____CAUAGGUGUAGAUCCAGUGGCAGAUUUCGAUAAGUCAUUGGGCUAUAAUUUGC ............((((((...(((....)))(((((((.(((......))).)))))))))).)))(((((((......((((....))))..))))))) (-24.89 = -24.86 + -0.03)

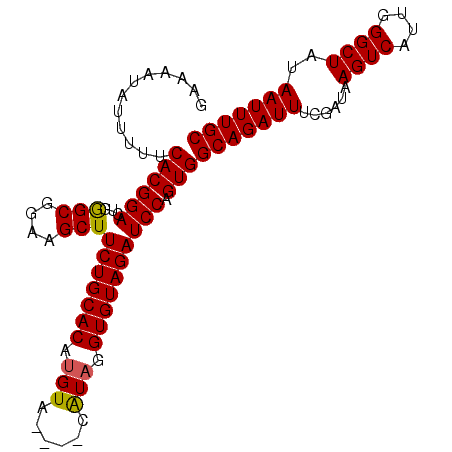

| Location | 12,188,546 – 12,188,651 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 95.05 |
| Mean single sequence MFE | -15.30 |
| Consensus MFE | -12.25 |
| Energy contribution | -13.05 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.561542 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12188546 105 + 27905053 UACUGUCGACGAGAAAUUUUUUCAUUAUAAUUAGACACAUCACAGGGAAACAACAACAAUUUCCACAAAAACCGACUAAAAAUAAAGUUGACAUUUCACACAGCA ...(((((((((((.....)))).((((..((((...........(((((.........)))))...........))))..)))).)))))))............ ( -14.45) >DroSec_CAF1 2346 105 + 1 UACUGUCGACGUGAAAUUUUUUCAUUAUACUUAGACACAUCACAGGGAAACAACAACAAUUUCCACAAAAACCGACUACAAAUAAAGUUGACAUUUCACACAGCA ..((((....(((((((............................(((((.........)))))........(((((........)))))..))))))))))).. ( -14.30) >DroSim_CAF1 2345 105 + 1 UACUGUCGACGUGAAAUUUUUUCAUUAUAAUUAGACACAUCAGAGGGAAACAACAACAAUUUCCACAAAAACCGACUAAAAAUAAAGUUGACAUUUCACACAGCA ..((((....(((((((.((..(.((((..((((...........(((((.........)))))...........))))..)))).)..)).))))))))))).. ( -14.75) >DroEre_CAF1 2591 105 + 1 UACUGUCGACGUGAAAUUUUUUCAUUAUAAUUAGGCACAUCAUAGGGAAACAGCAACAGCUUCCACAAAAACCGACUAAAAAUAAAGUUGACAUUUCACACAGCG ..((((....(((((((.((..(.((((..((((...........(((...(((....))))))...........))))..)))).)..)).))))))))))).. ( -17.55) >DroYak_CAF1 2289 105 + 1 UACUGUCGACGUGAAAUUUUUUCAUUAUAAUUAGGCACAUCAUAGAGAAACAACAACAGUUUCCACAAAAACCGACUAAACAUAAAGUUGACAUUUCACACAGCA ..((((....(((((((.((..(.((((..((((............(((((.......)))))............))))..)))).)..)).))))))))))).. ( -15.45) >consensus UACUGUCGACGUGAAAUUUUUUCAUUAUAAUUAGACACAUCACAGGGAAACAACAACAAUUUCCACAAAAACCGACUAAAAAUAAAGUUGACAUUUCACACAGCA ..((((....(((((((.((..(.((((..((((...........(((((.........)))))...........))))..)))).)..)).))))))))))).. (-12.25 = -13.05 + 0.80)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:15 2006