
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
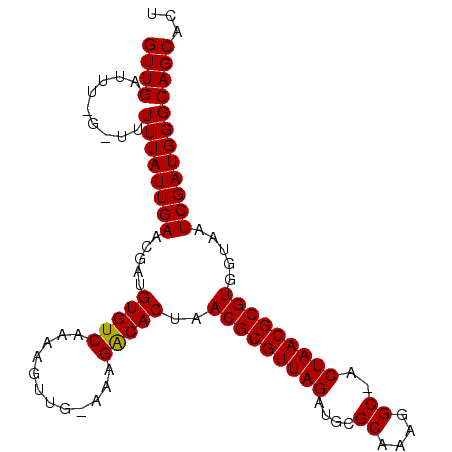
| Location | 12,184,992 – 12,185,114 |
| Length | 122 |
| Max. P | 0.982825 |

| Location | 12,184,992 – 12,185,090 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 92.97 |
| Mean single sequence MFE | -28.52 |
| Consensus MFE | -24.12 |
| Energy contribution | -23.88 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.936884 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
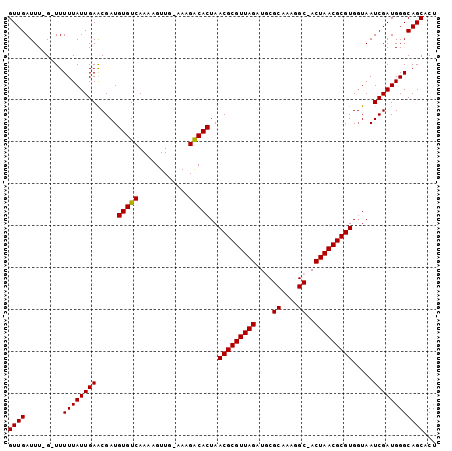
>3R_DroMel_CAF1 12184992 98 + 27905053 GUUGAUUU-G-UUUUUAUUGAACGAUGUGUCAAAAGUUG-GAAGACACUAACGCGUUAGAUGCGCACAGGC-ACUAACGCGUGGUAAUCGAUGGGCAGCACU .......(-(-((((((((((.....(((((........-...)))))..(((((((((.(((......))-)))))))))).....)))))))).)))).. ( -29.60) >DroSec_CAF1 9530 98 + 1 GUUGAUUU-G-UUUUUAUUGAACGAUGUGUCAAAAGUUG-AAUGACACUAACGCGUUAGAUGCGCAAAGGC-ACUAACGCGUGGUAAUCGAUGGGCAGCACU .......(-(-((((((((((.....((((((.......-..))))))..(((((((((.(((......))-)))))))))).....)))))))).)))).. ( -30.70) >DroSim_CAF1 10247 101 + 1 GUUGAUUUUGUUUUUUAUUGAACGAUGUGUCAAACGUUGGAAUGACACUAACGCGUUAGAUGCGCACAGGCAACUAACGCGUGGUAAUCGAUGGGCAGC-CU .........(((.((((((((.((((((.....)))))).......((((.((((((((.(((......))).))))))))))))..)))))))).)))-.. ( -31.20) >DroEre_CAF1 8975 98 + 1 GUUGAUUU-G-UUUUUAUUGAACGAUGUGUCAAAUGUUG-AAAGGCACUAACGCGUUAGAUACGCAAAAGC-ACUAACGCGUGGUAAUCGAUGGGCAGCACU ......((-(-(((.....)))))).((((....(((((-(...((....(((((((((....((....))-.))))))))).))..))))))....)))). ( -25.20) >DroYak_CAF1 9537 98 + 1 GUUGAUUU-G-UUUUUAUUGAGCGAUGUGUCAAAAGUUG-AAAGGCACUAACGCGUUAGAUACGCAAACGC-ACUAACGCGUGUUAAUCGAUGGGCAGCACU .......(-(-((((((((((..((((((((........-...)))))..(((((((((....((....))-.))))))))))))..)))))))).)))).. ( -25.90) >consensus GUUGAUUU_G_UUUUUAUUGAACGAUGUGUCAAAAGUUG_AAAGACACUAACGCGUUAGAUGCGCAAAGGC_ACUAACGCGUGGUAAUCGAUGGGCAGCACU ((((.........((((((((.....(((((............)))))..(((((((((....((....))..))))))))).....))))))))))))... (-24.12 = -23.88 + -0.24)

| Location | 12,184,992 – 12,185,090 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 92.97 |
| Mean single sequence MFE | -23.89 |
| Consensus MFE | -19.48 |
| Energy contribution | -19.88 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.982825 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12184992 98 - 27905053 AGUGCUGCCCAUCGAUUACCACGCGUUAGU-GCCUGUGCGCAUCUAACGCGUUAGUGUCUUC-CAACUUUUGACACAUCGUUCAAUAAAAA-C-AAAUCAAC .((.........((((....((((((((((-((......))).)))))))))..(((((...-........)))))))))..........)-)-........ ( -25.11) >DroSec_CAF1 9530 98 - 1 AGUGCUGCCCAUCGAUUACCACGCGUUAGU-GCCUUUGCGCAUCUAACGCGUUAGUGUCAUU-CAACUUUUGACACAUCGUUCAAUAAAAA-C-AAAUCAAC .((.........((((....((((((((((-((......))).)))))))))..((((((..-.......))))))))))..........)-)-........ ( -26.51) >DroSim_CAF1 10247 101 - 1 AG-GCUGCCCAUCGAUUACCACGCGUUAGUUGCCUGUGCGCAUCUAACGCGUUAGUGUCAUUCCAACGUUUGACACAUCGUUCAAUAAAAAACAAAAUCAAC .(-(....))...((((...(((((((((.(((......))).)))))))))..((((((..(....)..))))))...................))))... ( -27.60) >DroEre_CAF1 8975 98 - 1 AGUGCUGCCCAUCGAUUACCACGCGUUAGU-GCUUUUGCGUAUCUAACGCGUUAGUGCCUUU-CAACAUUUGACACAUCGUUCAAUAAAAA-C-AAAUCAAC .((.........((((....((((((((((-((......))).)))))))))..(((....(-((.....))))))))))..........)-)-........ ( -18.71) >DroYak_CAF1 9537 98 - 1 AGUGCUGCCCAUCGAUUAACACGCGUUAGU-GCGUUUGCGUAUCUAACGCGUUAGUGCCUUU-CAACUUUUGACACAUCGCUCAAUAAAAA-C-AAAUCAAC .(.((...............((((((((((-(((....)))).)))))))))..(((....(-((.....))))))...)).)........-.-........ ( -21.50) >consensus AGUGCUGCCCAUCGAUUACCACGCGUUAGU_GCCUUUGCGCAUCUAACGCGUUAGUGUCUUU_CAACUUUUGACACAUCGUUCAAUAAAAA_C_AAAUCAAC .............((((...(((((((((..((....))....)))))))))..(((((............)))))...................))))... (-19.48 = -19.88 + 0.40)

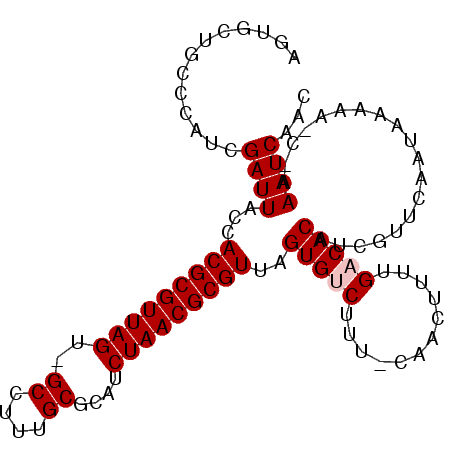

| Location | 12,185,013 – 12,185,114 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 91.55 |
| Mean single sequence MFE | -32.24 |
| Consensus MFE | -24.96 |
| Energy contribution | -24.72 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947478 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12185013 101 + 27905053 GAUGUGUCAAAAGUUG-GAAGACACUAACGCGUUAGAUGCGCACAGGC-ACUAACGCGUGGUAAUCGAUGGGCAGCACUGGUGUGCUCAAAAUGAGCACAGCU ...((((.....((((-(...((....(((((((((.(((......))-)))))))))).))..))))).....)))).(.(((((((.....))))))).). ( -38.30) >DroSec_CAF1 9551 101 + 1 GAUGUGUCAAAAGUUG-AAUGACACUAACGCGUUAGAUGCGCAAAGGC-ACUAACGCGUGGUAAUCGAUGGGCAGCACUGGUGUGCUCAAAAUGAGCAGAACU ...((((.....((((-(...((....(((((((((.(((......))-)))))))))).))..))))).....)))).(((.(((((.....)))))..))) ( -34.40) >DroSim_CAF1 10270 102 + 1 GAUGUGUCAAACGUUGGAAUGACACUAACGCGUUAGAUGCGCACAGGCAACUAACGCGUGGUAAUCGAUGGGCAGC-CUGGUGUGGUCAAAAUGAGCAGAACU ...........((((...((.(((((((((((((((.(((......))).)))))))))(((..((....))..))-))))))).))...))))......... ( -30.20) >DroEre_CAF1 8996 101 + 1 GAUGUGUCAAAUGUUG-AAAGGCACUAACGCGUUAGAUACGCAAAAGC-ACUAACGCGUGGUAAUCGAUGGGCAGCACUGGUGAGCUCAAAAUGAGCAGAGUU ...((((....(((((-(...((....(((((((((....((....))-.))))))))).))..))))))....))))......((((.....))))...... ( -28.60) >DroYak_CAF1 9558 101 + 1 GAUGUGUCAAAAGUUG-AAAGGCACUAACGCGUUAGAUACGCAAACGC-ACUAACGCGUGUUAAUCGAUGGGCAGCACUGGUGAGCUCAAAAUGAGCUGAGCU ...((((.....((((-(..(((((....(((((((....((....))-.))))))))))))..))))).....)))).(.(.(((((.....))))).).). ( -29.70) >consensus GAUGUGUCAAAAGUUG_AAAGACACUAACGCGUUAGAUGCGCAAAGGC_ACUAACGCGUGGUAAUCGAUGGGCAGCACUGGUGUGCUCAAAAUGAGCAGAGCU ((((((((............)))))..(((((((((....((....))..)))))))))....))).............(.(.(((((.....))))).).). (-24.96 = -24.72 + -0.24)



| Location | 12,185,013 – 12,185,114 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 91.55 |
| Mean single sequence MFE | -28.06 |
| Consensus MFE | -21.66 |
| Energy contribution | -22.22 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.951865 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12185013 101 - 27905053 AGCUGUGCUCAUUUUGAGCACACCAGUGCUGCCCAUCGAUUACCACGCGUUAGU-GCCUGUGCGCAUCUAACGCGUUAGUGUCUUC-CAACUUUUGACACAUC ((((((((((.....))))))).....)))..............((((((((((-((......))).)))))))))..(((((...-........)))))... ( -34.10) >DroSec_CAF1 9551 101 - 1 AGUUCUGCUCAUUUUGAGCACACCAGUGCUGCCCAUCGAUUACCACGCGUUAGU-GCCUUUGCGCAUCUAACGCGUUAGUGUCAUU-CAACUUUUGACACAUC .....(((((.....)))))........................((((((((((-((......))).)))))))))..((((((..-.......))))))... ( -30.10) >DroSim_CAF1 10270 102 - 1 AGUUCUGCUCAUUUUGACCACACCAG-GCUGCCCAUCGAUUACCACGCGUUAGUUGCCUGUGCGCAUCUAACGCGUUAGUGUCAUUCCAACGUUUGACACAUC .....((..((.((((.......)))-).))..)).........(((((((((.(((......))).)))))))))..((((((..(....)..))))))... ( -25.50) >DroEre_CAF1 8996 101 - 1 AACUCUGCUCAUUUUGAGCUCACCAGUGCUGCCCAUCGAUUACCACGCGUUAGU-GCUUUUGCGUAUCUAACGCGUUAGUGCCUUU-CAACAUUUGACACAUC ......((((.....))))......(..(((.............((((((((((-((......))).))))))))))))..)....-................ ( -22.91) >DroYak_CAF1 9558 101 - 1 AGCUCAGCUCAUUUUGAGCUCACCAGUGCUGCCCAUCGAUUAACACGCGUUAGU-GCGUUUGCGUAUCUAACGCGUUAGUGCCUUU-CAACUUUUGACACAUC (((((((......))))))).....(..(((.............((((((((((-(((....)))).))))))))))))..)....-................ ( -27.71) >consensus AGCUCUGCUCAUUUUGAGCACACCAGUGCUGCCCAUCGAUUACCACGCGUUAGU_GCCUUUGCGCAUCUAACGCGUUAGUGUCUUU_CAACUUUUGACACAUC .....(((((.....)))))........................(((((((((..((....))....)))))))))..(((((............)))))... (-21.66 = -22.22 + 0.56)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:12 2006