
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,179,258 – 12,179,360 |
| Length | 102 |
| Max. P | 0.583701 |

| Location | 12,179,258 – 12,179,360 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 81.12 |
| Mean single sequence MFE | -29.33 |
| Consensus MFE | -16.03 |
| Energy contribution | -17.78 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.583701 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
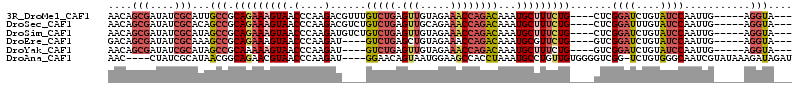
>3R_DroMel_CAF1 12179258 102 - 27905053 AACAGCGAUAUCGCAUUGCCGCAGAAAGUAACCCAAGACGUUUGUCUGAGUUGUAGAAACCAGACAAAUGCUUUCUG----CUCGGAUCUGUAUCCAAUUG-----AGGUA--- ....(((....)))..(((((((((((((..........(((((((((.(((.....))))))))))))))))))))----)..((((....)))).....-----.))))--- ( -35.70) >DroSec_CAF1 3964 102 - 1 AACAGCGAUAUCGCACAGCCGCAGAAAGUAACCCAAGACGUCUGUCUGAGUUGCAGAAACCAGACAAAUGCUUUCUG----CUCGGAUUUGUAUCCAAUUG-----AGGUA--- ....(((....)))...(((((((((((((.....((....))(((((.(((.....))))))))...)))))))))----)..((((....)))).....-----.))).--- ( -34.70) >DroSim_CAF1 4020 102 - 1 AACAGCGAUAUCGCAUAGCCGCAGAAAGUAACCCAAGAUGUCUGUCUGAGUUGUAGAAACCAGACAAAUGCUUUCUG----CUCGGAUCUGUAUCCAAUUG-----AGGUA--- ....(((....)))...(((((((((((((.....((....))(((((.(((.....))))))))...)))))))))----)..((((....)))).....-----.))).--- ( -33.90) >DroEre_CAF1 3813 98 - 1 GACAGCGAUAUCGCAAAGCCGCAGAAAGUAACCCAAGAU----GUCUGAGCUGUAGAAACCAGACAAAUGCGUUCUG----GUCGGAUCUGUAUCCAAUUG-----AGGUA--- ....(((....)))...(((.(((((.(((.(....).(----(((((............))))))..))).)))))----...((((....)))).....-----.))).--- ( -21.70) >DroYak_CAF1 3929 98 - 1 AACAGCGAUAUCGCAUAGCCGCAAAAAGUAACCCAAGAU----GUCUGAGUUGUAGAAACCAGACAAAUGCUUUCUG----GUCGGAUCUGUAUCCAAUUG-----AGGUA--- ....(((....)))...(((.(((......(((.(((.(----(((((.(((.....)))))))))....)))...)----)).((((....))))..)))-----.))).--- ( -24.20) >DroAna_CAF1 4358 105 - 1 AAC----CUAUCGCAUAACGGCAGAACGUAACCCAAGAU----GGAACAGUAAUGGAAGCCACCUAAAUGCCUGUUGUGGGGUCGG-UCUGUGGGCAAUCGUAUAAAGAUAGAU ...----(((((......(.(((((.((...((((.(((----((..((.....((......))....)).))))).))))..)).-))))).)((....)).....))))).. ( -25.80) >consensus AACAGCGAUAUCGCAUAGCCGCAGAAAGUAACCCAAGAU____GUCUGAGUUGUAGAAACCAGACAAAUGCUUUCUG____CUCGGAUCUGUAUCCAAUUG_____AGGUA___ ....(((....)))...(((.(((((((((.(....)......(((((.(((.....))))))))...))))))))).......((((....))))...........))).... (-16.03 = -17.78 + 1.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:05 2006