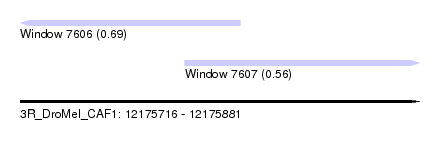
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,175,716 – 12,175,881 |
| Length | 165 |
| Max. P | 0.689491 |

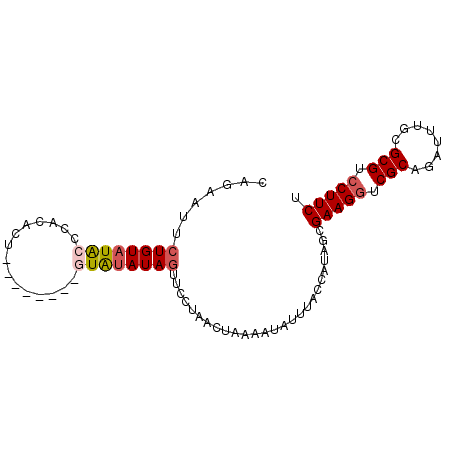
| Location | 12,175,716 – 12,175,807 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 81.49 |
| Mean single sequence MFE | -15.93 |
| Consensus MFE | -12.35 |
| Energy contribution | -13.16 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.689491 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12175716 91 - 27905053 UUGAAUUCUGUAUACCCACACUUCCUAUAUAUGUAUAGUUCCUAACUAAAAUAUUUACCAUAGCGAAGGUCGCAGAUUUGUGCGUCCUUCU ..(((..(((((((.................)))))))))).......................(((((.((((......)))).))))). ( -15.63) >DroSec_CAF1 484 83 - 1 CAGAAUUCUGUAUACCCACACU--------GUAUAUAGUCACUAACUAAAAUAUUUACCAUAGCGAAGGUCGCAGAUUUGCGCGUCCUUCU .......((((((((.......--------))))))))..........................(((((.(((........))).))))). ( -18.60) >DroSim_CAF1 485 83 - 1 CAGAAUUCUGUAUACCCACACU--------GUACAUAGUUCCUAACUAAAAUAUUUACCAUAGCGAAGGUCGCAGAUUUGCGCGUCCUUCU ..(((((.((((((.......)--------))))).))))).......................(((((.(((........))).))))). ( -19.50) >DroYak_CAF1 481 76 - 1 CAGAACACUGUAUG---------------UUUGUAUAUAUCCUAACUAAAAUAUUUACCAUAACGAAGAUCGCAGAUUUGCGCGUCCUUCU (((....)))...(---------------((.(((.((((..........)))).)))...)))((((..(((........)))..)))). ( -10.00) >consensus CAGAAUUCUGUAUACCCACACU________GUAUAUAGUUCCUAACUAAAAUAUUUACCAUAGCGAAGGUCGCAGAUUUGCGCGUCCUUCU .......((((((((...............))))))))..........................(((((.(((........))).))))). (-12.35 = -13.16 + 0.81)

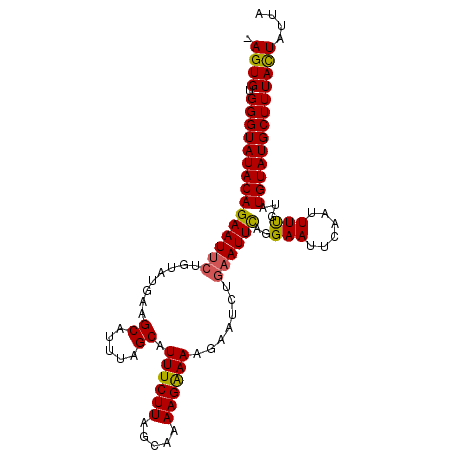
| Location | 12,175,784 – 12,175,881 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 85.71 |
| Mean single sequence MFE | -21.70 |
| Consensus MFE | -17.01 |
| Energy contribution | -16.80 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.556393 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12175784 97 + 27905053 AAGUGUGGGUAUACAGAAUUCAAUAUGAAGCACCUAGCAUUUCUUGGCAAAAGGAAAGA------AUUCAGGAAUACAAUUUCGCAUGUAUGCUUUACUAUUA .((((.(((((((((((((((........((.....)).((((((......))))))))------))))..(((......)))...))))))))))))).... ( -23.60) >DroSec_CAF1 545 102 + 1 -AGUGUGGGUAUACAGAAUUCUGUAUGAAGCAUUUAGCAUUUCUUAGCAAAAGAAAAGAAUCUGAAUUCAGGAAUUCAAUUUUGUAUGUAUGCUUUAUUAUUA -(((((...(((((((....)))))))..))))).((((((((((.....))))))(((((.((((((....))))))))))).......))))......... ( -21.40) >DroSim_CAF1 546 102 + 1 -AGUGUGGGUAUACAGAAUUCUGUAUGAAGCAUUUAGCAUUUCUUAGCCAAAGAAAAGAAUCUGAAUUUAGGAACUAAAUUUUUUAUGUAUGCUUUACUAUUA -((((.(((((((((((.((((.......((.....)).((((((.....)))))))))))))((((((((...)))))))).....)))))))))))).... ( -20.10) >consensus _AGUGUGGGUAUACAGAAUUCUGUAUGAAGCAUUUAGCAUUUCUUAGCAAAAGAAAAGAAUCUGAAUUCAGGAAUUCAAUUUUGUAUGUAUGCUUUACUAUUA .((((.(((((((((((((((........((.....)).((((((.....)))))).......))))))..(((......)))...))))))))))))).... (-17.01 = -16.80 + -0.21)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:02 2006