| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,169,404 – 12,169,499 |
| Length | 95 |
| Max. P | 0.867267 |

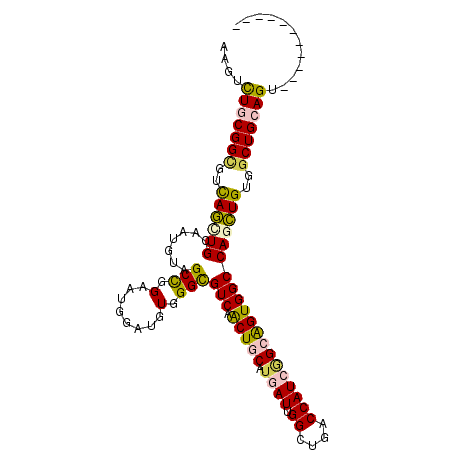
| Location | 12,169,404 – 12,169,499 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 79.66 |
| Mean single sequence MFE | -27.51 |
| Consensus MFE | -14.70 |
| Energy contribution | -15.20 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602194 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12169404 95 + 27905053 -----------ACUUCAGCCUCAGCUGGCCACUGCGGAUGGCCAGCCAAUCAUGCAGCUGACACCAACAUCAAUUCCGGCUACUCUGCAACUGACACCGCAAACAU -----------...((((.....(((((((((....).))))))))......(((((......((............)).....))))).))))............ ( -23.70) >DroPse_CAF1 85174 95 + 1 -----------ACUGCAGCCACAGCUGGCCACCGCCGAUGGUCAGCCAAUCAUGGAGUUGACGCCCACAUCCAUUCCAGCCACACUACAGCUGACGCCGCAGACUU -----------.((((.((..((((((......((.(((((....)).))).((((((.((........)).)))))))).......))))))..)).)))).... ( -29.32) >DroGri_CAF1 102961 106 + 1 UUCAAUUUGCAGCUGCAGCCACAAUUGGCCACUGCUGAUGGUCAGCCAAUAAUGCAGUUGACGCCGACUUCCAUUCCAGCCACAUUGCAGUUGACGCCGCAAACUU .....(((((.((..((((..((((((((....((((.....))))....((((.(((((....)))))..))))...)))).))))..))))..)).)))))... ( -29.70) >DroEre_CAF1 78804 95 + 1 -----------ACUUCAGCCUCAACUGGCCACUGCCGAUGGCCAGCCAAUCAUGCAGCUGACACCAACAUCAAUUCCGGCUACAUUGCAAUUGACACCGCAGACUC -----------...(((((..((.(((((((.......))))))).......))..)))))........((((((...((......))))))))............ ( -19.70) >DroAna_CAF1 76224 95 + 1 -----------GCUGCAGCCACAGUUGGCCACCGCUGAUGGACAGCCGAUAAUGCAGCUGACACCCACUUCAAUACCGGCUACUUUGCAGCUAACUCCGCAAACAU -----------((((((((((((((.(....).)))).)))..(((((.((.((.((.((.....)))).)).)).)))))...)))))))............... ( -27.70) >DroPer_CAF1 83174 95 + 1 -----------ACUGCAGCCACAGCUGGCCACCGCCGAUGGUCAGCCAAUCAUGGAGUUGACGCCCACGUCCAUUCCAGCCACACUACAGCUGACGCCGCAGACCU -----------.((((.((..((((((......((.(((((....)).))).((((((.((((....)))).)))))))).......))))))..)).)))).... ( -34.92) >consensus ___________ACUGCAGCCACAGCUGGCCACCGCCGAUGGUCAGCCAAUCAUGCAGCUGACACCCACAUCAAUUCCAGCCACACUGCAGCUGACGCCGCAAACUU ...............((((..((((((((((.......))))))))......))..))))........................((((.((....)).)))).... (-14.70 = -15.20 + 0.50)

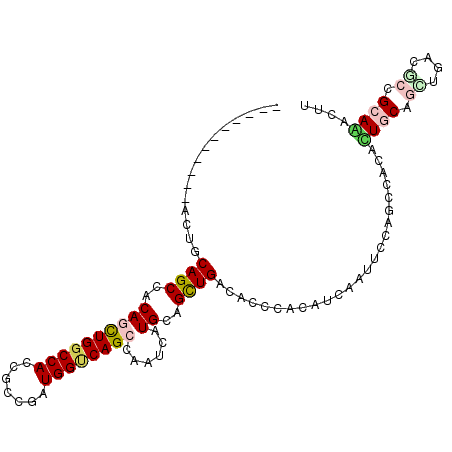

| Location | 12,169,404 – 12,169,499 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 79.66 |
| Mean single sequence MFE | -37.18 |
| Consensus MFE | -25.71 |
| Energy contribution | -25.10 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.867267 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12169404 95 - 27905053 AUGUUUGCGGUGUCAGUUGCAGAGUAGCCGGAAUUGAUGUUGGUGUCAGCUGCAUGAUUGGCUGGCCAUCCGCAGUGGCCAGCUGAGGCUGAAGU----------- .......((((.((...(((((....(((((........))))).....))))).....((((((((((.....)))))))))))).))))....----------- ( -36.40) >DroPse_CAF1 85174 95 - 1 AAGUCUGCGGCGUCAGCUGUAGUGUGGCUGGAAUGGAUGUGGGCGUCAACUCCAUGAUUGGCUGACCAUCGGCGGUGGCCAGCUGUGGCUGCAGU----------- ....(((((((..((((((....((.((((((...((((....))))...))).......((((.....))))))).))))))))..))))))).----------- ( -38.40) >DroGri_CAF1 102961 106 - 1 AAGUUUGCGGCGUCAACUGCAAUGUGGCUGGAAUGGAAGUCGGCGUCAACUGCAUUAUUGGCUGACCAUCAGCAGUGGCCAAUUGUGGCUGCAGCUGCAAAUUGAA .((((((((((..((.(..((((.(((((....(((..((((((..(((........)))))))))..))).....)))))))))..).))..))))))))))... ( -37.70) >DroEre_CAF1 78804 95 - 1 GAGUCUGCGGUGUCAAUUGCAAUGUAGCCGGAAUUGAUGUUGGUGUCAGCUGCAUGAUUGGCUGGCCAUCGGCAGUGGCCAGUUGAGGCUGAAGU----------- .((((((((((....))))))(((((((.(..(((......)))..).)))))))..(..(((((((((.....)))))))))..))))).....----------- ( -35.80) >DroAna_CAF1 76224 95 - 1 AUGUUUGCGGAGUUAGCUGCAAAGUAGCCGGUAUUGAAGUGGGUGUCAGCUGCAUUAUCGGCUGUCCAUCAGCGGUGGCCAACUGUGGCUGCAGC----------- ...(((((((......))))))).(((((((((.((.(((........))).)).))))))))).......((.(..((((....))))..).))----------- ( -34.60) >DroPer_CAF1 83174 95 - 1 AGGUCUGCGGCGUCAGCUGUAGUGUGGCUGGAAUGGACGUGGGCGUCAACUCCAUGAUUGGCUGACCAUCGGCGGUGGCCAGCUGUGGCUGCAGU----------- ....(((((((..((((((....((.((((((...((((....))))...))).......((((.....))))))).))))))))..))))))).----------- ( -40.20) >consensus AAGUCUGCGGCGUCAGCUGCAAUGUAGCCGGAAUGGAUGUGGGCGUCAACUGCAUGAUUGGCUGACCAUCGGCAGUGGCCAGCUGUGGCUGCAGU___________ ....(((((((..((((((.......(((.(........).)))(((.(((((.((((.((....))))))))))))))))))))..)))))))............ (-25.71 = -25.10 + -0.61)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:58 2006