| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,742,458 – 1,742,549 |
| Length | 91 |
| Max. P | 0.654275 |

| Location | 1,742,458 – 1,742,549 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 79.96 |
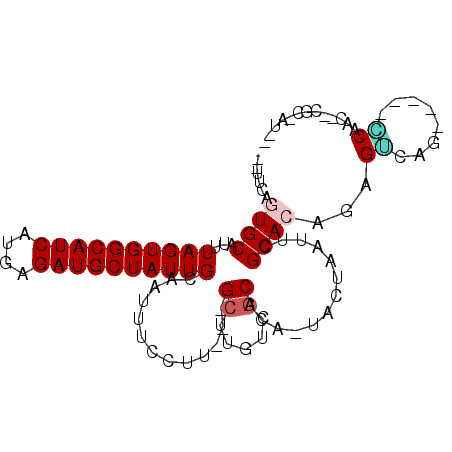
| Mean single sequence MFE | -23.52 |
| Consensus MFE | -16.11 |
| Energy contribution | -16.33 |
| Covariance contribution | 0.22 |
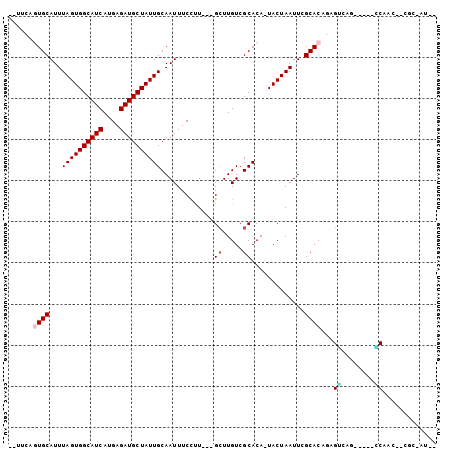
| Combinations/Pair | 1.17 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654275 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
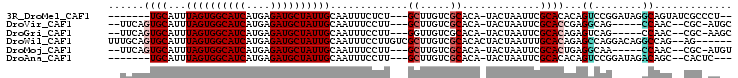
>3R_DroMel_CAF1 1742458 91 + 27905053 -------UGCAUUUAGUGGCAUCAUGAGAUGCUAUUGCAAUUUCUCU---GCUUGUCGCACA-UACUAAUUCGCACACAGUCCGGAUAGGCAGUAUCGCCCU-- -------.((...((((((((((....))))))))))........((---(((((((((...-.........))...(.....))))))))))....))...-- ( -23.60) >DroVir_CAF1 61301 90 + 1 --UUCAGUGCAUUUAGUGGCAUCAUGAGAUGCUAUUGCAAUUUCCUU---GCUUGUCGCACA-UACUAAUUCGCACCGAGGCAG-----CCAAC--CGC-AUGC --(((.((((....(((((((((....)))))))))((((.....))---))..........-.........)))).)))((((-----(....--.))-.))) ( -23.20) >DroGri_CAF1 53643 90 + 1 --UUCAGUGCAUUUAGUGGCAUCAUGAGAUGCUAUUGCAAUUUCCUU---GGUUGUCGCACA-UACUAAUUCGCACAGAGUCAG-----CCAAC--CGC-AAGC --....((((....(((((((((....)))))))))((((((.....---)))))).)))).-..((.((((.....)))).))-----.....--(..-..). ( -22.40) >DroWil_CAF1 45942 96 + 1 UUUGCAGUGCAUUUAGUGGCAUCAUGAGAUGCUAUUGCAAUUUCCUUGUCGCUUGUCGCACACUACUAAUUUGCACAGAGCCAGGACAGGCCAG--AG------ .(((((((((((((.(((....))).)))))).)))))))....((((((((((...(((...........)))...))))...))))))....--..------ ( -26.20) >DroMoj_CAF1 57478 90 + 1 --UUCAGUGCAUUUAGUGGCAUCAUGAGAUGCUAUUGCAAUUUCCUU---GCUUGUCGCACA-UACUAAUUCGCACUGAGGCAA-----CCAAC--CGC-AUGU --((((((((....(((((((((....)))))))))((((.....))---))..........-.........))))))))((..-----.....--.))-.... ( -26.30) >DroAna_CAF1 48006 88 + 1 -------UGCAUUUAGUGGCAUCAUGAGAUGCUAUUGCAAUUUCCUU---GCUUGUCGCACA-UACUAAUUCGCACACAGUCCGGAUAGACAGC--CACUC--- -------((((....((((((((....))))))))))))...(((..---((((((.((...-.........))))).)))..)))........--.....--- ( -19.40) >consensus __UUCAGUGCAUUUAGUGGCAUCAUGAGAUGCUAUUGCAAUUUCCUU___GCUUGUCGCACA_UACUAAUUCGCACAGAGUCAG_____CCAAC__CGC_AU__ ......((((...((((((((((....)))))))))).............((.....)).............))))...((........))............. (-16.11 = -16.33 + 0.22)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:48 2006