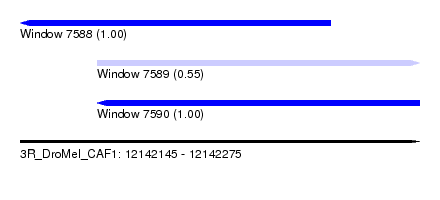
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,142,145 – 12,142,275 |
| Length | 130 |
| Max. P | 0.998741 |

| Location | 12,142,145 – 12,142,246 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 94.58 |
| Mean single sequence MFE | -25.63 |
| Consensus MFE | -22.70 |
| Energy contribution | -22.90 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.21 |
| SVM RNA-class probability | 0.998741 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12142145 101 - 27905053 CACUCUCAAGUGGCUCAUUGAGUUGCCUGCCAGCCAUCCAUAUACCAAUUCAUCUUGCAUCCACCGCCGGAUGUUGGGCAUCUC------CAUCUAUCAGCUAUUUG ......(((((((((.((.(((.((((((....)......................((((((......)))))).))))).)))------.....)).))))))))) ( -24.20) >DroSec_CAF1 48527 101 - 1 CACUCUCAAGUGGCUCAUUGAGUUGGCUUCCAGCCAUCCAUAUACCAAUUCAUCUUGCAUCCACCGCCGGAUGUUCGGCAUCUC------CAUCUAUCAGCUAUUUG ......(((((((((...((((((((..................))))))))((..((((((......))))))..))......------........))))))))) ( -24.17) >DroSim_CAF1 48841 101 - 1 CACUCUCAAGUGGCUCAUUGAGUUGGCUUCCAGCCAUCCAUAUACCAAUUCAUCUUGCAUCCACCGCCGGAUGUUCGGCAUCUC------CAUCUAUCAGCUAUUUG ......(((((((((...((((((((..................))))))))((..((((((......))))))..))......------........))))))))) ( -24.17) >DroEre_CAF1 48281 101 - 1 CACUCUCAAGUGGCUCAUUGAGUUGGCUUCUAGCCAUCCAUGUACCAAUUCAUCUUGCAUCCACCGCCGGAUGUUGGGCAUCUC------CAUCUAUCAGCUAUUUG ......(((((((((.((.((..((((.....))))...((((.((((..(((((.((.......)).)))))))))))))...------..)).)).))))))))) ( -27.50) >DroYak_CAF1 60015 107 - 1 CACUCUCAAGUGGCUCAUUGAGUUGGCUUCUAGCCAUCCAUAUAGCAAUGCAUCUUGCAUCCACCGCCGGAUGUUGGGCAUCUCCAUCUCCAUCUAUCAGCUAUUUG ......(((((((((.((.((..((((.....))))........((...((((((.((.......)).))))))...)).............)).)).))))))))) ( -28.10) >consensus CACUCUCAAGUGGCUCAUUGAGUUGGCUUCCAGCCAUCCAUAUACCAAUUCAUCUUGCAUCCACCGCCGGAUGUUGGGCAUCUC______CAUCUAUCAGCUAUUUG ......(((((((((.((.((..((((.....))))................((..((((((......))))))..))..............)).)).))))))))) (-22.70 = -22.90 + 0.20)

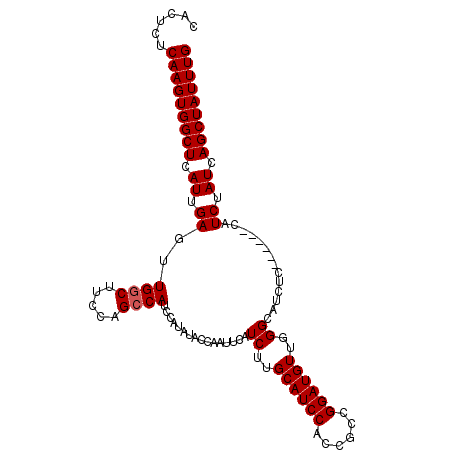
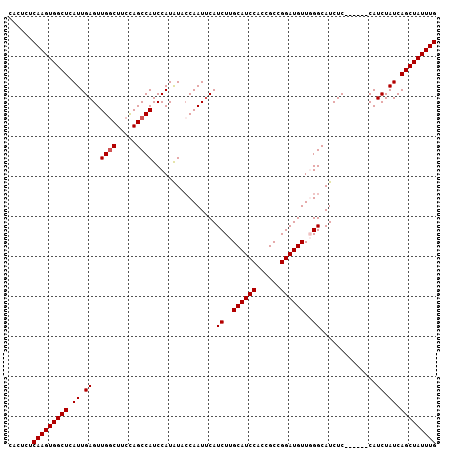
| Location | 12,142,170 – 12,142,275 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 94.06 |
| Mean single sequence MFE | -30.35 |
| Consensus MFE | -25.66 |
| Energy contribution | -26.10 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.548396 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12142170 105 + 27905053 CAACAUCCGGCGGUGGAUGCAAGAUGAAUUGGUAUAUGGAUGGCUGGCAGGCAACUCAAUGAGCCACUUGAGAGUGGGACAAUAGAGAAAGCCCCCCCCACUCUA ...((((..(((.....)))..))))..............(((((.(.((....))...).)))))....((((((((..................)))))))). ( -28.47) >DroSec_CAF1 48552 104 + 1 GAACAUCCGGCGGUGGAUGCAAGAUGAAUUGGUAUAUGGAUGGCUGGAAGCCAACUCAAUGAGCCACUUGAGAGUGGCACAAUAGAGAAAGCAC-CCCCACUCCA ........((.(((((.(((....................((((.....)))).(((..((.((((((....)))))).))...)))...))).-..))))))). ( -30.40) >DroSim_CAF1 48866 104 + 1 GAACAUCCGGCGGUGGAUGCAAGAUGAAUUGGUAUAUGGAUGGCUGGAAGCCAACUCAAUGAGCCACUUGAGAGUGGCACAAUAGAGAAAGCAC-CCCCACUCCA ........((.(((((.(((....................((((.....)))).(((..((.((((((....)))))).))...)))...))).-..))))))). ( -30.40) >DroEre_CAF1 48306 103 + 1 CAACAUCCGGCGGUGGAUGCAAGAUGAAUUGGUACAUGGAUGGCUAGAAGCCAACUCAAUGAGCCACUUGAGAGUGGCACAAUAGAGAAAGC-C-UCCCAAUCCA ...((((..(((.....)))..)))).(((((.....((.((((.....)))).(((..((.((((((....)))))).))...)))....)-)-..)))))... ( -29.70) >DroYak_CAF1 60046 103 + 1 CAACAUCCGGCGGUGGAUGCAAGAUGCAUUGCUAUAUGGAUGGCUAGAAGCCAACUCAAUGAGCCACUUGAGAGUGGCACAAUAGAGAAAGC-C-UCCCACUCCA ........((.(((((((((.....)))).(((.......((((.....)))).(((..((.((((((....)))))).))...)))..)))-.-..))))))). ( -32.80) >consensus CAACAUCCGGCGGUGGAUGCAAGAUGAAUUGGUAUAUGGAUGGCUGGAAGCCAACUCAAUGAGCCACUUGAGAGUGGCACAAUAGAGAAAGC_C_CCCCACUCCA ...((((..(((.....)))..))))...(((....(((.((((.....)))).(((..((.((((((....)))))).))...)))..........)))..))) (-25.66 = -26.10 + 0.44)

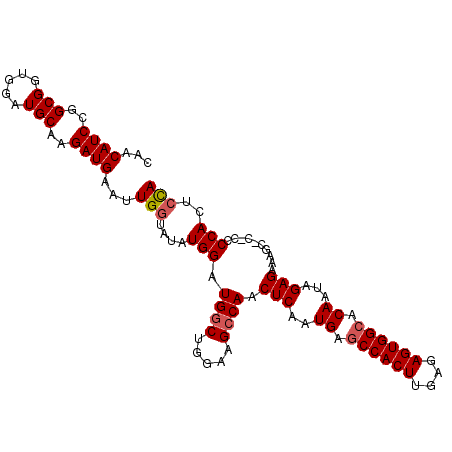

| Location | 12,142,170 – 12,142,275 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 94.06 |
| Mean single sequence MFE | -37.30 |
| Consensus MFE | -30.00 |
| Energy contribution | -30.12 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.995799 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12142170 105 - 27905053 UAGAGUGGGGGGGGCUUUCUCUAUUGUCCCACUCUCAAGUGGCUCAUUGAGUUGCCUGCCAGCCAUCCAUAUACCAAUUCAUCUUGCAUCCACCGCCGGAUGUUG ..(..(((((.((((...(((...((..(((((....)))))..))..)))..)))).))..)))..).................((((((......)))))).. ( -33.70) >DroSec_CAF1 48552 104 - 1 UGGAGUGGGG-GUGCUUUCUCUAUUGUGCCACUCUCAAGUGGCUCAUUGAGUUGGCUUCCAGCCAUCCAUAUACCAAUUCAUCUUGCAUCCACCGCCGGAUGUUC ((((.(((.(-(.(((..(((...((.((((((....)))))).))..)))..)))..))..)))))))................((((((......)))))).. ( -37.40) >DroSim_CAF1 48866 104 - 1 UGGAGUGGGG-GUGCUUUCUCUAUUGUGCCACUCUCAAGUGGCUCAUUGAGUUGGCUUCCAGCCAUCCAUAUACCAAUUCAUCUUGCAUCCACCGCCGGAUGUUC ((((.(((.(-(.(((..(((...((.((((((....)))))).))..)))..)))..))..)))))))................((((((......)))))).. ( -37.40) >DroEre_CAF1 48306 103 - 1 UGGAUUGGGA-G-GCUUUCUCUAUUGUGCCACUCUCAAGUGGCUCAUUGAGUUGGCUUCUAGCCAUCCAUGUACCAAUUCAUCUUGCAUCCACCGCCGGAUGUUG ((((((((((-(-(((..(((...((.((((((....)))))).))..)))..))))))..((.......)).))))))))....((((((......)))))).. ( -39.30) >DroYak_CAF1 60046 103 - 1 UGGAGUGGGA-G-GCUUUCUCUAUUGUGCCACUCUCAAGUGGCUCAUUGAGUUGGCUUCUAGCCAUCCAUAUAGCAAUGCAUCUUGCAUCCACCGCCGGAUGUUG ((((((.(((-(-(((..(((...((.((((((....)))))).))..)))..))))))).))..)))).........((((((.((.......)).)))))).. ( -38.70) >consensus UGGAGUGGGG_G_GCUUUCUCUAUUGUGCCACUCUCAAGUGGCUCAUUGAGUUGGCUUCCAGCCAUCCAUAUACCAAUUCAUCUUGCAUCCACCGCCGGAUGUUG ((((.(((.(...(((..(((...((.((((((....)))))).))..)))..)))...)..)))))))................((((((......)))))).. (-30.00 = -30.12 + 0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:46 2006