| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,128,610 – 12,128,702 |
| Length | 92 |
| Max. P | 0.991500 |

| Location | 12,128,610 – 12,128,702 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.64 |
| Mean single sequence MFE | -25.00 |
| Consensus MFE | -7.94 |
| Energy contribution | -8.14 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.32 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991500 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12128610 92 + 27905053 UUGACUUUUGUUUG------------------------GGAGCCUAAAAAUUAAUAGUUUCAGAGAAAUCCUAAAUUUUCG--CUAGGGGAAAAAUAAAAAGCGAAAAUAUGGAAGUC-- ..((((((..((((------------------------(((..((..((((.....))))...))...))))))(((((((--((...............))))))))))..))))))-- ( -20.76) >DroSec_CAF1 34919 116 + 1 UUGACUUUUGUUUGAGAAUGUUGAGAAUAGAAUAGUCUGGAGACUAAAAAUUAAUAUUUUCAGAGAAAUCCAAAAUUUUCG--CUAGGGGAAAAAUAAAAAGCGAAAAUAUGGAAGUC-- ..(((((((.((((((((((((((........(((((....)))))....)))))))))))))).)))((((..(((((((--((...............))))))))).))))))))-- ( -31.66) >DroSim_CAF1 34667 113 + 1 UUGACUUUUGUUUGAGAAUA---GGACCAACACGACCUGGAGACUAAAAAUUAAUAUUUUCAGAGAAAUCCUAAAUUUUCG--CUAGGGGAAAAAUAAAGAGCGAAAAUAUGGAAGUC-- ..(((((((.((((((((((---((.(......).)).(....)..........)))))))))).)))(((...(((((((--((...............)))))))))..)))))))-- ( -24.26) >DroEre_CAF1 35021 113 + 1 UUGACUUUUAUUUGGGAGUA---UAACCAACGCAA-CUGGAUACUGAAAAUUAAUAUUUUCUGAGAAAUCCUAAAGUUUUACCCUAGGGGGAAAAUGA-AAGCGAAACUAUGGAAGUC-- ..(((((((((((((.....---...))))(((..-..((((...((((((....))))))......))))....(((((.(((....))))))))..-..))).....)))))))))-- ( -26.20) >DroYak_CAF1 39291 113 + 1 UUGACUUUUAUUUGGGUAUA---UAACCAACACAA-CUGGAUACUGAAA--UAAUAUUUUUAGAGAAAUCCUAAAGUUUCGCCCUAGGGGAAAUCUGA-AAGUGAAACUAUGGAAUUAUA .......((((((.(((((.---...(((......-.)))))))).)))--)))((((((((((....(((((..(.....)..)))))....)))))-)))))................ ( -22.10) >consensus UUGACUUUUGUUUGAGAAUA___UAACCAACACAA_CUGGAGACUAAAAAUUAAUAUUUUCAGAGAAAUCCUAAAUUUUCG__CUAGGGGAAAAAUAAAAAGCGAAAAUAUGGAAGUC__ ..(((((((((...........................(((..((.(((((....)))))...))...)))...(((((((.....................)))))))))))))))).. ( -7.94 = -8.14 + 0.20)

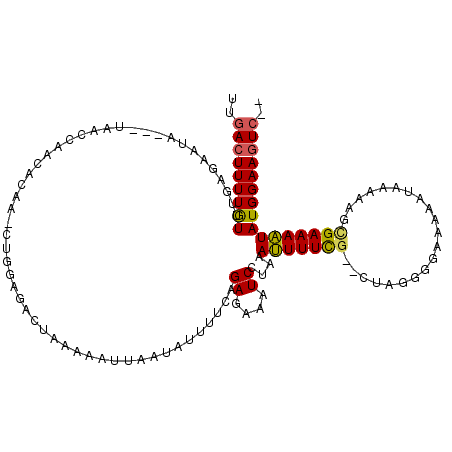

| Location | 12,128,610 – 12,128,702 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.64 |
| Mean single sequence MFE | -19.66 |
| Consensus MFE | -8.22 |
| Energy contribution | -9.30 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.42 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12128610 92 - 27905053 --GACUUCCAUAUUUUCGCUUUUUAUUUUUCCCCUAG--CGAAAAUUUAGGAUUUCUCUGAAACUAUUAAUUUUUAGGCUCC------------------------CAAACAAAAGUCAA --(((((((..(((((((((...............))--)))))))...)))....(((((((........)))))))....------------------------........)))).. ( -17.76) >DroSec_CAF1 34919 116 - 1 --GACUUCCAUAUUUUCGCUUUUUAUUUUUCCCCUAG--CGAAAAUUUUGGAUUUCUCUGAAAAUAUUAAUUUUUAGUCUCCAGACUAUUCUAUUCUCAACAUUCUCAAACAAAAGUCAA --((((((((.(((((((((...............))--)))))))..))))......(((.((((........(((((....)))))...)))).)))...............)))).. ( -21.86) >DroSim_CAF1 34667 113 - 1 --GACUUCCAUAUUUUCGCUCUUUAUUUUUCCCCUAG--CGAAAAUUUAGGAUUUCUCUGAAAAUAUUAAUUUUUAGUCUCCAGGUCGUGUUGGUCC---UAUUCUCAAACAAAAGUCAA --(((((....(((((((((...............))--))))))).(((((.....((((((((....))))))))...((((......)))))))---))...........))))).. ( -22.06) >DroEre_CAF1 35021 113 - 1 --GACUUCCAUAGUUUCGCUU-UCAUUUUCCCCCUAGGGUAAAACUUUAGGAUUUCUCAGAAAAUAUUAAUUUUCAGUAUCCAG-UUGCGUUGGUUA---UACUCCCAAAUAAAAGUCAA --(((((.........(((..-..........(((((((.....)))))))........((((((....)))))).........-..)))((((...---.....))))....))))).. ( -20.50) >DroYak_CAF1 39291 113 - 1 UAUAAUUCCAUAGUUUCACUU-UCAGAUUUCCCCUAGGGCGAAACUUUAGGAUUUCUCUAAAAAUAUUA--UUUCAGUAUCCAG-UUGUGUUGGUUA---UAUACCCAAAUAAAAGUCAA ......(((..((((((.(((-(.((.......)))))).))))))...)))..............(((--(((..((((((((-.....))))...---.))))..))))))....... ( -16.10) >consensus __GACUUCCAUAUUUUCGCUUUUUAUUUUUCCCCUAG__CGAAAAUUUAGGAUUUCUCUGAAAAUAUUAAUUUUUAGUCUCCAG_UUGUGUUGGUCA___UAUUCCCAAACAAAAGUCAA ..(((((((..(((((((.....................)))))))...))).....((((((((....)))))))).....................................)))).. ( -8.22 = -9.30 + 1.08)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:40 2006