
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
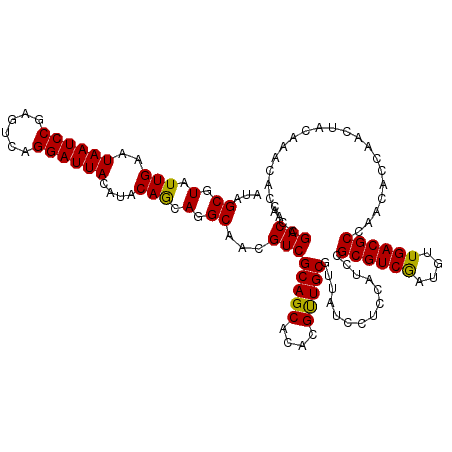
| Location | 12,127,672 – 12,127,821 |
| Length | 149 |
| Max. P | 0.999630 |

| Location | 12,127,672 – 12,127,792 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.22 |
| Mean single sequence MFE | -29.14 |
| Consensus MFE | -28.70 |
| Energy contribution | -28.12 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.98 |
| SVM decision value | 3.31 |
| SVM RNA-class probability | 0.998983 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12127672 120 + 27905053 AUAGCGUAUUGAAUAAUCCGAGUCAGGAUUAUAUCCAGCAGGCAACGUCGCAGCAUACGUUGCGUUAUCCUCCAUCCGCGUCGAUGUUGACGCCAACACCAACUACAAACACACCGACAC ...((.(.(((.(((((((......)))))))...))).).))...((((((((....)))))..............((((((....))))))......................))).. ( -28.90) >DroVir_CAF1 40973 120 + 1 AUAGCGUAUUGAAUAAUCCGAGUCAGGAUUACAUACAACAGGCAACGUCGCAGCACACGUUGCGUUAUCCUCCAUCCGCGUCGAUGUUGACGCCAACACCAACUACAAAUACACCGACAC ...((.(.(((..((((((......))))))....))).).))...((((((((....)))))..............((((((....))))))......................))).. ( -27.80) >DroPse_CAF1 38663 120 + 1 AUAGCGUAUUGAAUAAUCCGAGUCAGGAUUACAUCCAGCAGGCAACGUCGCAGCACACGCUGCGUUAUCCUCCAUCCGCGUCGAUGCUGACGCCCACACCAACUACAAACACACCGACAC ...((((......((((((......))))))....(((((..(.((((((((((....)))))).............)))).).)))))))))........................... ( -30.61) >DroGri_CAF1 44275 120 + 1 AUAGCGUAUUGAAUAAUCCGAGUCAGGAUUACAUACAACAAGCAACGUCGCAGCACACGUUGCGUUAUCCUCCAUCCGCGUCGAUGUUGACGCCAACACCAACUACAAAUACACCGACAC (((((((((....((((((......)))))).)))).....(((((((........)))))))))))).........((((((....))))))........................... ( -27.40) >DroWil_CAF1 39734 120 + 1 AUAGCGUAUUGAAUAAUCCAAGUCAGGAUUAUAUACAGCAGGCGACGUCGCAGCAUACGUUGCGUUAUCCUCCAUCCGCGUCAAUGUUGACGCCAACACCAACUACAAAUACACCGACAC ...((((((...(((((((......))))))))))).)).(....)((((((((....)))))..............((((((....))))))......................))).. ( -29.50) >DroPer_CAF1 37104 120 + 1 AUAGCGUAUUGAAUAAUCCGAGUCAGGAUUACAUCCAGCAGGCAACGUCGCAGCACACGCUGCGUUAUCCUCCAUCCGCGUCGAUGCUGACGCCCACACCAACUACAAACACACCGACAC ...((((......((((((......))))))....(((((..(.((((((((((....)))))).............)))).).)))))))))........................... ( -30.61) >consensus AUAGCGUAUUGAAUAAUCCGAGUCAGGAUUACAUACAGCAGGCAACGUCGCAGCACACGUUGCGUUAUCCUCCAUCCGCGUCGAUGUUGACGCCAACACCAACUACAAACACACCGACAC ...((.(.(((..((((((......))))))....))).).))...((((((((....)))))..............((((((....))))))......................))).. (-28.70 = -28.12 + -0.58)


| Location | 12,127,672 – 12,127,792 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.22 |
| Mean single sequence MFE | -40.17 |
| Consensus MFE | -36.83 |
| Energy contribution | -35.92 |
| Covariance contribution | -0.91 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12127672 120 - 27905053 GUGUCGGUGUGUUUGUAGUUGGUGUUGGCGUCAACAUCGACGCGGAUGGAGGAUAACGCAACGUAUGCUGCGACGUUGCCUGCUGGAUAUAAUCCUGACUCGGAUUAUUCAAUACGCUAU .....((((((((....((((((((((....))))))))))((((.....(.....)(((((((........)))))))))))..((.(((((((......))))))))))))))))).. ( -40.00) >DroVir_CAF1 40973 120 - 1 GUGUCGGUGUAUUUGUAGUUGGUGUUGGCGUCAACAUCGACGCGGAUGGAGGAUAACGCAACGUGUGCUGCGACGUUGCCUGUUGUAUGUAAUCCUGACUCGGAUUAUUCAAUACGCUAU .....((((((((.((((((.(.(((.(((((......)))))((((.....((((((((((((........)))))))..))))).....)))).))).).))))))..)))))))).. ( -41.20) >DroPse_CAF1 38663 120 - 1 GUGUCGGUGUGUUUGUAGUUGGUGUGGGCGUCAGCAUCGACGCGGAUGGAGGAUAACGCAGCGUGUGCUGCGACGUUGCCUGCUGGAUGUAAUCCUGACUCGGAUUAUUCAAUACGCUAU .....((((((((.((((((.(.((..(((((......)))))......(((((..((((((....))))))((((..((....)))))).))))).)).).))))))..)))))))).. ( -40.40) >DroGri_CAF1 44275 120 - 1 GUGUCGGUGUAUUUGUAGUUGGUGUUGGCGUCAACAUCGACGCGGAUGGAGGAUAACGCAACGUGUGCUGCGACGUUGCUUGUUGUAUGUAAUCCUGACUCGGAUUAUUCAAUACGCUAU .....((((((((.((((((.(.(((.(((((......)))))((((.....((((((((((((........)))))))..))))).....)))).))).).))))))..)))))))).. ( -41.20) >DroWil_CAF1 39734 120 - 1 GUGUCGGUGUAUUUGUAGUUGGUGUUGGCGUCAACAUUGACGCGGAUGGAGGAUAACGCAACGUAUGCUGCGACGUCGCCUGCUGUAUAUAAUCCUGACUUGGAUUAUUCAAUACGCUAU .....((((((((..((((.((((((.((((((....)))))).)))....(((..((((........))))..)))))).))))...(((((((......)))))))..)))))))).. ( -37.80) >DroPer_CAF1 37104 120 - 1 GUGUCGGUGUGUUUGUAGUUGGUGUGGGCGUCAGCAUCGACGCGGAUGGAGGAUAACGCAGCGUGUGCUGCGACGUUGCCUGCUGGAUGUAAUCCUGACUCGGAUUAUUCAAUACGCUAU .....((((((((.((((((.(.((..(((((......)))))......(((((..((((((....))))))((((..((....)))))).))))).)).).))))))..)))))))).. ( -40.40) >consensus GUGUCGGUGUAUUUGUAGUUGGUGUUGGCGUCAACAUCGACGCGGAUGGAGGAUAACGCAACGUGUGCUGCGACGUUGCCUGCUGGAUGUAAUCCUGACUCGGAUUAUUCAAUACGCUAU .....((((((((....((((((((((....))))))))))((((.....(.....)(((((((........))))))))))).....(((((((......)))))))..)))))))).. (-36.83 = -35.92 + -0.91)



| Location | 12,127,712 – 12,127,821 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.32 |
| Mean single sequence MFE | -23.80 |
| Consensus MFE | -23.65 |
| Energy contribution | -23.15 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.99 |
| SVM decision value | 3.81 |
| SVM RNA-class probability | 0.999630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12127712 109 + 27905053 GGCAACGUCGCAGCAUACGUUGCGUUAUCCUCCAUCCGCGUCGAUGUUGACGCCAACACCAACUACAAACACACCGACACUUCCGUUGUAAUUUAU----AUAAAUAUAAA-GC------ .((((((.((((((....)))))).............((((((....))))))..............................))))))..(((((----(....))))))-..------ ( -23.00) >DroPse_CAF1 38703 115 + 1 GGCAACGUCGCAGCACACGCUGCGUUAUCCUCCAUCCGCGUCGAUGCUGACGCCCACACCAACUACAAACACACCGACACUUCCGUUGUAAUUUAU----AUAAAUAUAAA-ACAAACAC .((((((.((((((....)))))).............((((((....))))))..............................))))))..(((((----(....))))))-........ ( -25.60) >DroGri_CAF1 44315 114 + 1 AGCAACGUCGCAGCACACGUUGCGUUAUCCUCCAUCCGCGUCGAUGUUGACGCCAACACCAACUACAAAUACACCGACACUUCCGUUGUAAUUUAUAUAUAUAAAUACAGAAAC------ .((((((.((((((....)))))).............((((((....))))))..............................)))))).((((((....))))))........------ ( -22.80) >DroWil_CAF1 39774 109 + 1 GGCGACGUCGCAGCAUACGUUGCGUUAUCCUCCAUCCGCGUCAAUGUUGACGCCAACACCAACUACAAAUACACCGACACUUCCGUUGUAAUUUAU----AUAAAUAUAAA-AC------ .((((((.((((((....)))))).............((((((....))))))..............................))))))..(((((----(....))))))-..------ ( -23.40) >DroAna_CAF1 29648 108 + 1 GGCAACGUCGCAGCACACGUUGCGUUAUCCUCCAUCCGCGUCGAUGCUGACGCCCACACCAACUACAAACACACCGACACUUCCGUUGUAAUUAA-----AAGAUUAAAAA-AA------ .((((((.((((((....)))))).............((((((....))))))..............................))))))......-----...........-..------ ( -22.40) >DroPer_CAF1 37144 115 + 1 GGCAACGUCGCAGCACACGCUGCGUUAUCCUCCAUCCGCGUCGAUGCUGACGCCCACACCAACUACAAACACACCGACACUUCCGUUGUAAUUUAU----AUUAAUAUAAA-ACAAACAC .((((((.((((((....)))))).............((((((....))))))..............................))))))..(((((----(....))))))-........ ( -25.60) >consensus GGCAACGUCGCAGCACACGUUGCGUUAUCCUCCAUCCGCGUCGAUGCUGACGCCAACACCAACUACAAACACACCGACACUUCCGUUGUAAUUUAU____AUAAAUAUAAA_AC______ .((((((.((((((....)))))).............((((((....))))))..............................))))))............................... (-23.65 = -23.15 + -0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:38 2006