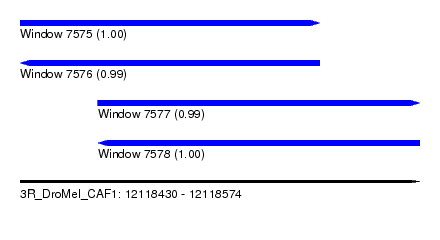
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,118,430 – 12,118,574 |
| Length | 144 |
| Max. P | 0.999735 |

| Location | 12,118,430 – 12,118,538 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 89.82 |
| Mean single sequence MFE | -23.25 |
| Consensus MFE | -22.63 |
| Energy contribution | -22.38 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.85 |
| SVM RNA-class probability | 0.997407 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12118430 108 + 27905053 UUUCUU-----------UUAUUUCAGGCAAAAACCUUUACAACAAUGAACAUGUAGCAAUAAAAAUGGAGCCUAUGAAGUCAAAGGCUCCGCAACUACACUUAGAGUAUAGGUUUUAUA ......-----------............(((((((.(((..........................(((((((.((....)).)))))))....(((....))).))).)))))))... ( -24.90) >DroVir_CAF1 30023 108 + 1 UCUUUU-----------CUUUUGCAGGAAAAAAUCUUUACAACAAUGAACAUGUAGCAAUAAAAAUGGAGCCUAUGAAGUCAAAGGCUCCGCAACUACACUUAGAGUAUAGGUUUUAUA ....((-----------((......))))(((((((.(((..........................(((((((.((....)).)))))))....(((....))).))).)))))))... ( -23.40) >DroGri_CAF1 32513 108 + 1 UUCUCU-----------CUUUUGCAGGAAAAAAUCUUUACAACAAUGAACAUGUAGCAAUAAAAAUGGAGCCGAUGAAGUCAAAGGCUCCGCAACUACACUUAGAGUAUAGGUUUUAUA ((((..-----------(....)..))))(((((((.(((..........................((((((..((....))..))))))....(((....))).))).)))))))... ( -21.50) >DroWil_CAF1 28978 112 + 1 UUUC---AAUUUC----UUUUUACAGGCAAAAAUCUUUACAACAAUGAACAUGUAGCAAUAAAAAUGGAGCCUAUGAAGUCAAAGGCUCCGCAACUACACUUAGAGUAUAGGUUUUAUA ....---......----............(((((((.(((..........................(((((((.((....)).)))))))....(((....))).))).)))))))... ( -22.30) >DroMoj_CAF1 30010 108 + 1 CUCUUU-----------CUUUUGCAGGAAAAAAUCUUUACAACAAUGAACAUGUAGCAAUAAAAAUGGAGCCUAUGAAGUCGAAGGCUCCACAACUACACUUAGAGUAUAGGUUUUAUA ....((-----------((......))))(((((((.(((.........................((((((((.((....)).))))))))...(((....))).))).)))))))... ( -23.40) >DroAna_CAF1 20077 119 + 1 UUUAUUCACUUUCCUCUGUUUUUCAGGCAAAAAUCUUUACAACAAUGAACAUGUAGCAAUAAAAAUGGAGCCUAUGAAGUCAAAGGCUCCGCAACUACACUUAGAGUAUAGGUUUUAUA ..............((((.....))))..(((((((.(((..........................(((((((.((....)).)))))))....(((....))).))).)))))))... ( -24.00) >consensus UUUUUU___________CUUUUGCAGGAAAAAAUCUUUACAACAAUGAACAUGUAGCAAUAAAAAUGGAGCCUAUGAAGUCAAAGGCUCCGCAACUACACUUAGAGUAUAGGUUUUAUA .............................(((((((.(((.........................((((((((.((....)).))))))))...(((....))).))).)))))))... (-22.63 = -22.38 + -0.25)

| Location | 12,118,430 – 12,118,538 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 89.82 |
| Mean single sequence MFE | -27.93 |
| Consensus MFE | -24.53 |
| Energy contribution | -24.83 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.989001 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12118430 108 - 27905053 UAUAAAACCUAUACUCUAAGUGUAGUUGCGGAGCCUUUGACUUCAUAGGCUCCAUUUUUAUUGCUACAUGUUCAUUGUUGUAAAGGUUUUUGCCUGAAAUAA-----------AAGAAA ........((((((.....))))))....(((((((.((....)).))))))).((((((((((.(((.......))).))..((((....))))..)))))-----------)))... ( -28.70) >DroVir_CAF1 30023 108 - 1 UAUAAAACCUAUACUCUAAGUGUAGUUGCGGAGCCUUUGACUUCAUAGGCUCCAUUUUUAUUGCUACAUGUUCAUUGUUGUAAAGAUUUUUUCCUGCAAAAG-----------AAAAGA ........((((((.....))))))(((((((((((.((....)).)))))))((((((((....(((.......))).))))))))........))))...-----------...... ( -28.30) >DroGri_CAF1 32513 108 - 1 UAUAAAACCUAUACUCUAAGUGUAGUUGCGGAGCCUUUGACUUCAUCGGCUCCAUUUUUAUUGCUACAUGUUCAUUGUUGUAAAGAUUUUUUCCUGCAAAAG-----------AGAGAA .............((((...(((((....((((((..((....))..))))))((((((((....(((.......))).))))))))......)))))....-----------)))).. ( -28.00) >DroWil_CAF1 28978 112 - 1 UAUAAAACCUAUACUCUAAGUGUAGUUGCGGAGCCUUUGACUUCAUAGGCUCCAUUUUUAUUGCUACAUGUUCAUUGUUGUAAAGAUUUUUGCCUGUAAAAA----GAAAUU---GAAA ........((((((.....))))))(((((((((((.((....)).)))))))((((((((....(((.......))).))))))))........))))...----......---.... ( -26.30) >DroMoj_CAF1 30010 108 - 1 UAUAAAACCUAUACUCUAAGUGUAGUUGUGGAGCCUUCGACUUCAUAGGCUCCAUUUUUAUUGCUACAUGUUCAUUGUUGUAAAGAUUUUUUCCUGCAAAAG-----------AAAGAG .............((((..(((((((.(((((((((..(....)..))))))))).......))))))).(((....(((((..((.....)).)))))..)-----------)))))) ( -27.40) >DroAna_CAF1 20077 119 - 1 UAUAAAACCUAUACUCUAAGUGUAGUUGCGGAGCCUUUGACUUCAUAGGCUCCAUUUUUAUUGCUACAUGUUCAUUGUUGUAAAGAUUUUUGCCUGAAAAACAGAGGAAAGUGAAUAAA .((((((.((((((.....))))))....(((((((.((....)).)))))))..)))))).......((((((((...(((((....)))))(((.....))).....)))))))).. ( -28.90) >consensus UAUAAAACCUAUACUCUAAGUGUAGUUGCGGAGCCUUUGACUUCAUAGGCUCCAUUUUUAUUGCUACAUGUUCAUUGUUGUAAAGAUUUUUGCCUGCAAAAG___________AAAAAA ........((((((.....))))))(((((((((((.((....)).)))))))((((((((....(((.......))).))))))))........)))).................... (-24.53 = -24.83 + 0.31)


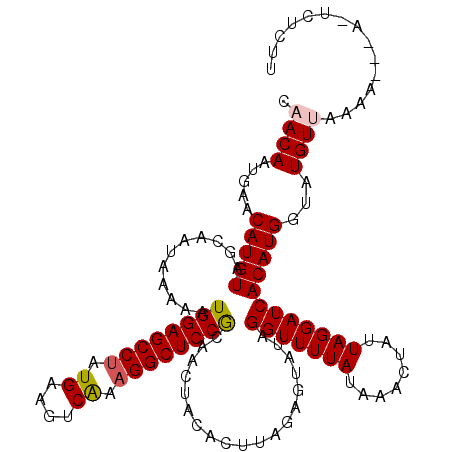
| Location | 12,118,458 – 12,118,574 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -26.87 |
| Consensus MFE | -24.19 |
| Energy contribution | -24.42 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.992569 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12118458 116 + 27905053 CAACAAUGAACAUGUAGCAAUAAAAAUGGAGCCUAUGAAGUCAAAGGCUCCGCAACUACACUUAGAGUAUAGGUUUUAUAAACUAUUAGGAUCACAUGGUAUGUAAU---CUA-UCGCUU ............(((((.........((((((((.((....)).))))))))...)))))....((((((((((..((((.(((((.........))))).))))))---)))-).)))) ( -25.10) >DroVir_CAF1 30051 115 + 1 CAACAAUGAACAUGUAGCAAUAAAAAUGGAGCCUAUGAAGUCAAAGGCUCCGCAACUACACUUAGAGUAUAGGUUUUAUAAGCUAUUAGGAUCACAUGGUAUGUUAAAU-----UGUCUU ..(((((.((((((((((.((((((..(((((((.((....)).)))))))....(((.((.....)).))).))))))..)))))....(((....))))))))..))-----)))... ( -27.60) >DroGri_CAF1 32541 119 + 1 CAACAAUGAACAUGUAGCAAUAAAAAUGGAGCCGAUGAAGUCAAAGGCUCCGCAACUACACUUAGAGUAUAGGUUUUAUAAGCUAUUAGGAUCACAUGGUAUGUUGAAU-UGAAUGUUUC .......((((((....((((......((((((..((....))..)))))).(((((((((.....))...(((((((........))))))).....))).)))).))-)).)))))). ( -27.60) >DroWil_CAF1 29010 114 + 1 CAACAAUGAACAUGUAGCAAUAAAAAUGGAGCCUAUGAAGUCAAAGGCUCCGCAACUACACUUAGAGUAUAGGUUUUAUAAACUAUUAGGAUCACAUGGUAUGUUGAAA-UUA-UC---- (((((.....(((((............(((((((.((....)).)))))))....................(((((((........))))))))))))...)))))...-...-..---- ( -28.10) >DroMoj_CAF1 30038 115 + 1 CAACAAUGAACAUGUAGCAAUAAAAAUGGAGCCUAUGAAGUCGAAGGCUCCACAACUACACUUAGAGUAUAGGUUUUAUAAGCUAUUAGGAUCACAUGGUAUGUUAAAA-----UGUUUU .((((...((((((((((.((((((.((((((((.((....)).))))))))...(((.((.....)).))).))))))..)))))....(((....))))))))....-----)))).. ( -26.70) >DroAna_CAF1 20116 119 + 1 CAACAAUGAACAUGUAGCAAUAAAAAUGGAGCCUAUGAAGUCAAAGGCUCCGCAACUACACUUAGAGUAUAGGUUUUAUAAACUAUUAGGAUCACAUGGUAUGUAAAAAAAAA-UCUCUU ............(((((.........((((((((.((....)).))))))))...)))))...((((......(((((((.(((((.........))))).))))))).....-.)))). ( -26.10) >consensus CAACAAUGAACAUGUAGCAAUAAAAAUGGAGCCUAUGAAGUCAAAGGCUCCGCAACUACACUUAGAGUAUAGGUUUUAUAAACUAUUAGGAUCACAUGGUAUGUUAAAA___A_UCUCUU .((((.....(((((...........((((((((.((....)).))))))))...................(((((((........))))))))))))...))))............... (-24.19 = -24.42 + 0.22)


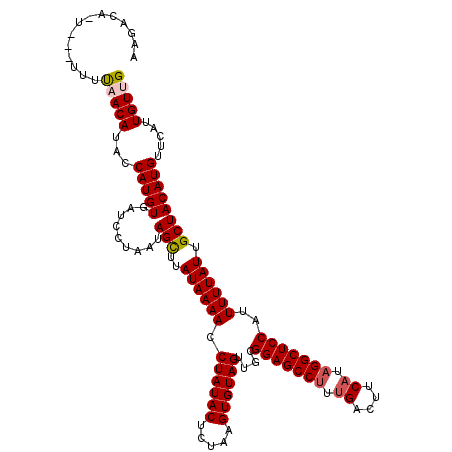
| Location | 12,118,458 – 12,118,574 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -29.85 |
| Consensus MFE | -27.27 |
| Energy contribution | -27.47 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.39 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.97 |
| SVM RNA-class probability | 0.999735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12118458 116 - 27905053 AAGCGA-UAG---AUUACAUACCAUGUGAUCCUAAUAGUUUAUAAAACCUAUACUCUAAGUGUAGUUGCGGAGCCUUUGACUUCAUAGGCUCCAUUUUUAUUGCUACAUGUUCAUUGUUG .(((((-(.(---(((((((...))))))))....((((..((((((.((((((.....))))))....(((((((.((....)).)))))))..)))))).)))).......)))))). ( -34.20) >DroVir_CAF1 30051 115 - 1 AAGACA-----AUUUAACAUACCAUGUGAUCCUAAUAGCUUAUAAAACCUAUACUCUAAGUGUAGUUGCGGAGCCUUUGACUUCAUAGGCUCCAUUUUUAUUGCUACAUGUUCAUUGUUG ..((((-----((..(((((..(....).......((((..((((((.((((((.....))))))....(((((((.((....)).)))))))..)))))).)))).))))).)))))). ( -30.70) >DroGri_CAF1 32541 119 - 1 GAAACAUUCA-AUUCAACAUACCAUGUGAUCCUAAUAGCUUAUAAAACCUAUACUCUAAGUGUAGUUGCGGAGCCUUUGACUUCAUCGGCUCCAUUUUUAUUGCUACAUGUUCAUUGUUG ..........-...(((((...(((((.........(((..((((((.((((((.....))))))....((((((..((....))..))))))..)))))).)))))))).....))))) ( -29.10) >DroWil_CAF1 29010 114 - 1 ----GA-UAA-UUUCAACAUACCAUGUGAUCCUAAUAGUUUAUAAAACCUAUACUCUAAGUGUAGUUGCGGAGCCUUUGACUUCAUAGGCUCCAUUUUUAUUGCUACAUGUUCAUUGUUG ----..-...-...(((((...((((((....((((((..........((((((.....))))))....(((((((.((....)).)))))))....)))))).)))))).....))))) ( -29.20) >DroMoj_CAF1 30038 115 - 1 AAAACA-----UUUUAACAUACCAUGUGAUCCUAAUAGCUUAUAAAACCUAUACUCUAAGUGUAGUUGUGGAGCCUUCGACUUCAUAGGCUCCAUUUUUAUUGCUACAUGUUCAUUGUUG ......-----...(((((...(((((.........(((..((((((.((((((.....))))))..(((((((((..(....)..))))))))))))))).)))))))).....))))) ( -28.40) >DroAna_CAF1 20116 119 - 1 AAGAGA-UUUUUUUUUACAUACCAUGUGAUCCUAAUAGUUUAUAAAACCUAUACUCUAAGUGUAGUUGCGGAGCCUUUGACUUCAUAGGCUCCAUUUUUAUUGCUACAUGUUCAUUGUUG ..(((.-.......((((((...))))))......((((..((((((.((((((.....))))))....(((((((.((....)).)))))))..)))))).))))....)))....... ( -27.50) >consensus AAGACA_U___UUUUAACAUACCAUGUGAUCCUAAUAGCUUAUAAAACCUAUACUCUAAGUGUAGUUGCGGAGCCUUUGACUUCAUAGGCUCCAUUUUUAUUGCUACAUGUUCAUUGUUG ..............(((((...(((((.........(((..((((((.((((((.....))))))....(((((((.((....)).)))))))..)))))).)))))))).....))))) (-27.27 = -27.47 + 0.19)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:34 2006