
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,109,515 – 12,109,676 |
| Length | 161 |
| Max. P | 0.999889 |

| Location | 12,109,515 – 12,109,623 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 72.73 |
| Mean single sequence MFE | -22.18 |
| Consensus MFE | -15.66 |
| Energy contribution | -16.11 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.976804 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
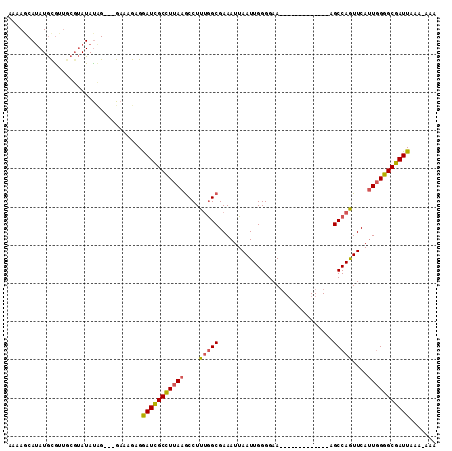
>3R_DroMel_CAF1 12109515 108 + 27905053 AAAAUCAUAUGCGUUGCGUAUAUCG---GAAAGAGGAUCGCUUUAAGCCUUAAGCGAAA-UAAUUGGGGAACGCCAGUUUUUUAAGCCAGUUCAAUUGGGCGAUUAAUUUGA ..(((((((((((...))))))).(---(........((((((........))))))..-...(((((((((....))))))))).)).((((....))))))))....... ( -22.10) >DroSec_CAF1 15592 95 + 1 AAAAGCAAAUGUAUUGCGUAUAUUG---GAAAGAGGAUCGCCUUAAGCCUUUGGCGAAAUUAAUUGGGGAA-------------AGCCAAUUCAUUGGGGCGAUUAAA-AAA ....((((.....))))........---.......((((((((((((((...)))......((((((....-------------..))))))..)))))))))))...-... ( -25.00) >DroSim_CAF1 15535 94 + 1 AAAAGCAUAUACGC----AAUACAGGAAGAAAGAGAAUUGCCUUAAACCUUUGGCGAAAUUAAUUGGGGAA-------------AGCCAGUUCAUUGGGGCGAUUAAA-AAA ....((......))----.................(((((((((((....(((((................-------------.)))))....)))))))))))...-... ( -19.43) >consensus AAAAGCAUAUGCGUUGCGUAUAUAG___GAAAGAGGAUCGCCUUAAGCCUUUGGCGAAAUUAAUUGGGGAA_____________AGCCAGUUCAUUGGGGCGAUUAAA_AAA ...................................(((((((((((....(((((..............................)))))....)))))))))))....... (-15.66 = -16.11 + 0.45)


| Location | 12,109,545 – 12,109,652 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 75.37 |
| Mean single sequence MFE | -26.89 |
| Consensus MFE | -18.89 |
| Energy contribution | -19.24 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.70 |
| SVM decision value | 4.40 |
| SVM RNA-class probability | 0.999889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12109545 107 + 27905053 AGGAUCGCUUUAAGCCUUAAGCGAAA-UAAUUGGGGAACGCCAGUUUUUUAAGCCAGUUCAAUUGGGCGAUUAAUUUGAAUUUGUACAAAAAGCGGG---------GUGUAAAUUAA ....((((((........))))))..-..........(((((.(((((((..((.(((((((((((....)))).))))))).))..)))))))..)---------))))....... ( -25.10) >DroSec_CAF1 15622 103 + 1 AGGAUCGCCUUAAGCCUUUGGCGAAAUUAAUUGGGGAA-------------AGCCAAUUCAUUGGGGCGAUUAAA-AAAAUUUGUACGAAAAGCGGGAAUAAAUGCGUACAAAUAAA ..((((((((((((((...)))......((((((....-------------..))))))..)))))))))))...-...(((((((((.................)))))))))... ( -29.83) >DroSim_CAF1 15564 103 + 1 AGAAUUGCCUUAAACCUUUGGCGAAAUUAAUUGGGGAA-------------AGCCAGUUCAUUGGGGCGAUUAAA-AAAAUUUGUACGAAAAGUGGGAACAAAUGCGUGUAAAUAAA ..(((((((((((....(((((................-------------.)))))....)))))))))))...-...(((((((((.....((....))....)))))))))... ( -25.73) >consensus AGGAUCGCCUUAAGCCUUUGGCGAAAUUAAUUGGGGAA_____________AGCCAGUUCAUUGGGGCGAUUAAA_AAAAUUUGUACGAAAAGCGGGAA_AAAUGCGUGUAAAUAAA ..(((((((((((....(((((..............................)))))....))))))))))).......(((((((((.................)))))))))... (-18.89 = -19.24 + 0.35)



| Location | 12,109,583 – 12,109,676 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 75.09 |
| Mean single sequence MFE | -18.88 |
| Consensus MFE | -12.32 |
| Energy contribution | -12.33 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981119 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
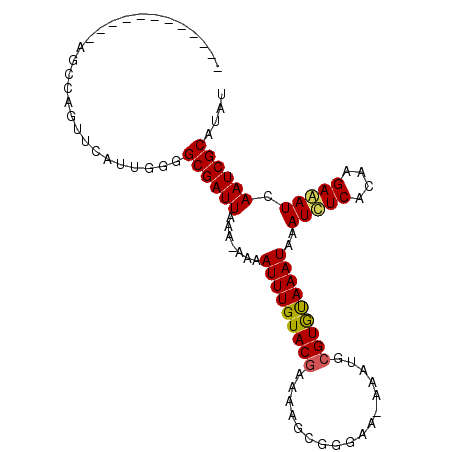
>3R_DroMel_CAF1 12109583 93 + 27905053 GCCAGUUUUUUAAGCCAGUUCAAUUGGGCGAUUAAUUUGAAUUUGUACAAAAAGCGGG---------GUGUAAAUUAAUCUCACAAGAGAUCAAUCGCAUAU .((.(((((((..((.(((((((((((....)))).))))))).))..))))))).))---------((((..(((.(((((....))))).))).)))).. ( -20.00) >DroSec_CAF1 15660 89 + 1 ------------AGCCAAUUCAUUGGGGCGAUUAAA-AAAAUUUGUACGAAAAGCGGGAAUAAAUGCGUACAAAUAAAUUUCACAAGAAAUCAAUCGCAUAU ------------..((((....)))).((((((...-...(((((((((.................)))))))))..(((((....))))).)))))).... ( -19.23) >DroSim_CAF1 15602 89 + 1 ------------AGCCAGUUCAUUGGGGCGAUUAAA-AAAAUUUGUACGAAAAGUGGGAACAAAUGCGUGUAAAUAAAUCUCAGAAGAAAUCAAUCGCAUAU ------------..((((....)))).((((((...-...(((((((((.....((....))....)))))))))...(((....)))....)))))).... ( -17.40) >consensus ____________AGCCAGUUCAUUGGGGCGAUUAAA_AAAAUUUGUACGAAAAGCGGGAA_AAAUGCGUGUAAAUAAAUCUCACAAGAAAUCAAUCGCAUAU ...........................((((((.......(((((((((.................)))))))))..(((((....))))).)))))).... (-12.32 = -12.33 + 0.01)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:27 2006