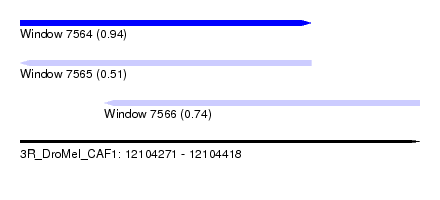
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,104,271 – 12,104,418 |
| Length | 147 |
| Max. P | 0.935796 |

| Location | 12,104,271 – 12,104,378 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 91.08 |
| Mean single sequence MFE | -24.79 |
| Consensus MFE | -19.96 |
| Energy contribution | -20.52 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12104271 107 + 27905053 --AAAAAAUUCUUGUCAAAUUUCUCGCUCCAGCUCUUGAAAUUUCAAGAGCAGCACCAUGAAGAAAAUGAAAAGGGACAAGUGCUGUUCAACUAACUCAGCUGGCUUAA --........((((((.........(((...((((((((....)))))))))))..(((.......)))......)))))).((((...........))))........ ( -25.20) >DroSec_CAF1 10255 109 + 1 CCUACAAAUUCUUGUCAAAUUUCUUGCUCCAGCUCUUGAAAUUUCAAGAGCAGCACCAUGAAGAAAAUGAAAAGGGACAAGUGCUGUUCAACUAACUCAGCUGGCUUAA .............((((.....((((..((.((((((((....)))))))).....(((.......)))....))..)))).((((...........)))))))).... ( -26.80) >DroSim_CAF1 10197 109 + 1 CCUACAAAUUCUUGUCAAAUUUCUUGCUCCAGCUCUUGAAAUUUCAAGAGCAGCACCAUGAAGAAAAUGAAAAGGGACAAGUGCUGUUCAACUAACUCAGCUGGCUUAA .............((((.....((((..((.((((((((....)))))))).....(((.......)))....))..)))).((((...........)))))))).... ( -26.80) >DroEre_CAF1 9918 103 + 1 ------AAUUCUUGUCAAAUUUCUCACUUCAGCUCUGGAAAUUUCAAGAGCAGAGCCAUGAAGAAAAUGAAAAGGGACAAGUGCUGUUCAACUAACUCGGCGUGCUUAA ------....((((((...((((...(((((((((((.............))))))..))))).....))))...)))))).((((...........))))........ ( -24.82) >DroYak_CAF1 14390 109 + 1 CAUUCAAAUUCUUGUCAAAUUUCUCACUUCAGCUCUGAAAAUUUCAAGAGCAGAGCCAUGAAGAAAAUGAAAAGGAACAAGUGCUGUUCAACUAACUCGGCUGGCUUAA .............((((..((((...(((((((((((.............))))))..))))).....))))..(((((.....)))))............)))).... ( -20.32) >consensus C_UACAAAUUCUUGUCAAAUUUCUCGCUCCAGCUCUUGAAAUUUCAAGAGCAGCACCAUGAAGAAAAUGAAAAGGGACAAGUGCUGUUCAACUAACUCAGCUGGCUUAA ..........((((((...............((((((((....)))))))).....(((.......)))......)))))).((((...........))))........ (-19.96 = -20.52 + 0.56)



| Location | 12,104,271 – 12,104,378 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 91.08 |
| Mean single sequence MFE | -25.97 |
| Consensus MFE | -23.11 |
| Energy contribution | -23.11 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.89 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.511626 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12104271 107 - 27905053 UUAAGCCAGCUGAGUUAGUUGAACAGCACUUGUCCCUUUUCAUUUUCUUCAUGGUGCUGCUCUUGAAAUUUCAAGAGCUGGAGCGAGAAAUUUGACAAGAAUUUUUU-- ........((((...........)))).((((((...((((....((.....))((((((((((((....))))))))...)))).))))...))))))........-- ( -26.80) >DroSec_CAF1 10255 109 - 1 UUAAGCCAGCUGAGUUAGUUGAACAGCACUUGUCCCUUUUCAUUUUCUUCAUGGUGCUGCUCUUGAAAUUUCAAGAGCUGGAGCAAGAAAUUUGACAAGAAUUUGUAGG .....((.((((...........)))).((((((...((((....((.....))((((((((((((....))))))))...)))).))))...))))))........)) ( -28.30) >DroSim_CAF1 10197 109 - 1 UUAAGCCAGCUGAGUUAGUUGAACAGCACUUGUCCCUUUUCAUUUUCUUCAUGGUGCUGCUCUUGAAAUUUCAAGAGCUGGAGCAAGAAAUUUGACAAGAAUUUGUAGG .....((.((((...........)))).((((((...((((....((.....))((((((((((((....))))))))...)))).))))...))))))........)) ( -28.30) >DroEre_CAF1 9918 103 - 1 UUAAGCACGCCGAGUUAGUUGAACAGCACUUGUCCCUUUUCAUUUUCUUCAUGGCUCUGCUCUUGAAAUUUCCAGAGCUGAAGUGAGAAAUUUGACAAGAAUU------ ........((...(((.....))).)).((((((..((((((((((......(((((((.............)))))))))))))))))....))))))....------ ( -22.62) >DroYak_CAF1 14390 109 - 1 UUAAGCCAGCCGAGUUAGUUGAACAGCACUUGUUCCUUUUCAUUUUCUUCAUGGCUCUGCUCUUGAAAUUUUCAGAGCUGAAGUGAGAAAUUUGACAAGAAUUUGAAUG ......((..((((((.(((....))).((((((..((((((((((......(((((((.............)))))))))))))))))....))))))))))))..)) ( -23.82) >consensus UUAAGCCAGCUGAGUUAGUUGAACAGCACUUGUCCCUUUUCAUUUUCUUCAUGGUGCUGCUCUUGAAAUUUCAAGAGCUGGAGCGAGAAAUUUGACAAGAAUUUGUA_G ........((((...........)))).((((((...((((.((..(((((.......((((((((....))))))))))))).))))))...)))))).......... (-23.11 = -23.11 + -0.00)



| Location | 12,104,302 – 12,104,418 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 91.90 |
| Mean single sequence MFE | -35.96 |
| Consensus MFE | -26.60 |
| Energy contribution | -27.60 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.743253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12104302 116 - 27905053 GGAGAUUCAAGGGGUGGCACGGAGGAAGAGAAAAGCGAACUUAAGCCAGCUGAGUUAGUUGAACAGCACUUGUCCCUUUUCAUUUUCUUCAUGGUGCUGCUCUUGAAAUUUCAAGA .((((((..(((((..((((.((((((((((((((.((...((((...((((...........)))).)))))).)))))).))))))))...))))..)))))..)))))).... ( -40.40) >DroSec_CAF1 10288 116 - 1 GGAGAUUCAAGGGGUGGCACCGAGGAAGAUAAAAGCGAACUUAAGCCAGCUGAGUUAGUUGAACAGCACUUGUCCCUUUUCAUUUUCUUCAUGGUGCUGCUCUUGAAAUUUCAAGA .((((((..(((((..(((((((((((((((((((.((...((((...((((...........)))).)))))).))))).)))))))))..)))))..)))))..)))))).... ( -40.40) >DroSim_CAF1 10230 116 - 1 GGAGAUUCAAGGGGUGGCACCGAGGAAGAUAAAAGCGAACUUAAGCCAGCUGAGUUAGUUGAACAGCACUUGUCCCUUUUCAUUUUCUUCAUGGUGCUGCUCUUGAAAUUUCAAGA .((((((..(((((..(((((((((((((((((((.((...((((...((((...........)))).)))))).))))).)))))))))..)))))..)))))..)))))).... ( -40.40) >DroEre_CAF1 9945 116 - 1 GGAGACUUAAGUGGUGGCAUGGUAGGAGAUAAAAGCGACCUUAAGCACGCCGAGUUAGUUGAACAGCACUUGUCCCUUUUCAUUUUCUUCAUGGCUCUGCUCUUGAAAUUUCCAGA (((((.((.((..(.(.(((((.((((((((((((.(((....(((.......))).(((....)))....))).))))).)))))))))))).).)..))....)).)))))... ( -28.40) >DroYak_CAF1 14423 116 - 1 GGAGACUCAAGUGGUGGCAUGGUAGGAGAUAAAAGCGAACUUAAGCCAGCCGAGUUAGUUGAACAGCACUUGUUCCUUUUCAUUUUCUUCAUGGCUCUGCUCUUGAAAUUUUCAGA (((((.(((((..(.(.(((((.((((((((((((.((((....((((((.......))))....))....))))))))).)))))))))))).).)..))..)))..)))))... ( -30.20) >consensus GGAGAUUCAAGGGGUGGCACGGAGGAAGAUAAAAGCGAACUUAAGCCAGCUGAGUUAGUUGAACAGCACUUGUCCCUUUUCAUUUUCUUCAUGGUGCUGCUCUUGAAAUUUCAAGA ((((.((((((.((..((((.((((((((((((((.((...((((...((((...........)))).)))))).))))).)))))))))...))))..)))))))).)))).... (-26.60 = -27.60 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:23 2006