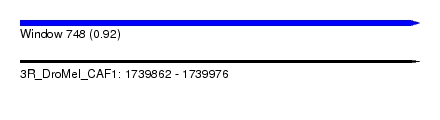
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,739,862 – 1,739,976 |
| Length | 114 |
| Max. P | 0.924665 |

| Location | 1,739,862 – 1,739,976 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.06 |
| Mean single sequence MFE | -22.48 |
| Consensus MFE | -12.80 |
| Energy contribution | -13.80 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924665 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
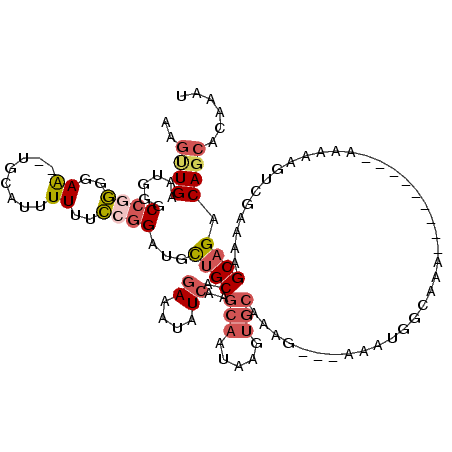
>3R_DroMel_CAF1 1739862 114 + 27905053 AAGUUGAAUGGGCCGGGGAA--UGCAUUUUUUCCGGAUGCUGCAGAAAUAUCAAGCAAUAAGUGCAAAG---AAAUGGCAAAAAAAAAAAAAAAACGUCAAAAAGCAGACAGCACAAAU ..((((......((((..((--......))..))))...((((...........(((.....)))....---...((((.................))))....)))).))))...... ( -23.93) >DroPse_CAF1 42347 98 + 1 CGACUGAAUAGGCCAAGGAGCCUUCUUUUUUUUGCGCUUCAGCACAAAAAUCAAGAAA----------AAUAAUAUACAAAA---------AAGUAACCG--AAGCAGACAACGUGAAU ...(((....(((......)))(((((.((((((.((....)).))))))..))))).----------..............---------.........--...)))........... ( -15.30) >DroSec_CAF1 48902 106 + 1 AAGUUGAAUGGGCCGGGGAA--UGCAUUUUUUCCGGAUGCUGCAGAAAUAUCAAGCAAUAAGUGCAAAG---AAAUGGCAAAA--------AAAACGUCGAAAAGCAGACAGCACAAAU ..((((......((((..((--......))..))))...((((...........(((.....)))....---...((((....--------.....))))....)))).))))...... ( -23.90) >DroSim_CAF1 50104 106 + 1 AAGUUGAAUGGGCCGGGGAA--UGCAUUUUUUCCGGAUGCUGCAGAAAUAUCAAGCAAUAAGUGCAAAG---AAAUGGCAAAA--------AAAACGUCGAAAAGCAGACAGCACAAAU ..((((......((((..((--......))..))))...((((...........(((.....)))....---...((((....--------.....))))....)))).))))...... ( -23.90) >DroEre_CAF1 47778 105 + 1 AAGCUGAAUGGGCCGGGGAA--UACAUUUUUUCCGGAUGCUGCAGAAAUAUCAAGCAAUAAGUGCAAAG---AAAUGGUAAA---------AAAAAGUCGAAGAGCAGACAGCAGAAAU ..((((......((((..((--......))..))))...((((.((....))..(((.....)))....---..........---------.............)))).))))...... ( -24.20) >DroYak_CAF1 49373 108 + 1 AAGUUGAAUGGGCCAGGGAA--UGCAUUUUUUCCGGAUGUUGCAGAAACAUCAAGCAAUAAGUGCAAAAAAAAAAUGCUAAA---------AAGAAGUCCAAAAGCAGACAGCACAAAU ..(((...(((((.......--.(((((((((...((((((.....))))))..(((.....)))....)))))))))....---------.....)))))..)))............. ( -23.63) >consensus AAGUUGAAUGGGCCGGGGAA__UGCAUUUUUUCCGGAUGCUGCAGAAAUAUCAAGCAAUAAGUGCAAAG___AAAUGGCAAA_________AAAAAGUCGAAAAGCAGACAGCACAAAU ..((((......((((..((........))..))))...((((.((....))..(((.....))).......................................)))).))))...... (-12.80 = -13.80 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:46 2006