
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,080,667 – 12,080,776 |
| Length | 109 |
| Max. P | 0.981722 |

| Location | 12,080,667 – 12,080,776 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.18 |
| Mean single sequence MFE | -35.62 |
| Consensus MFE | -30.16 |
| Energy contribution | -30.32 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.973065 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
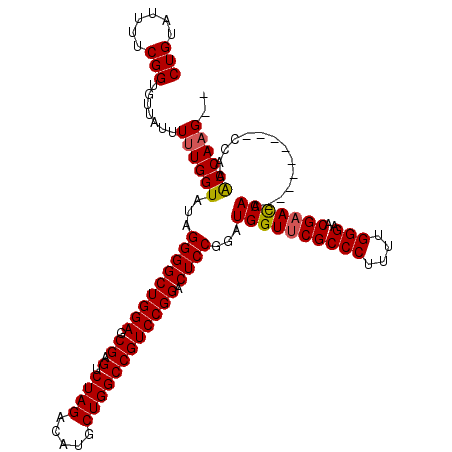
>3R_DroMel_CAF1 12080667 109 + 27905053 --CUUGGUUUGG---------UUGGUUCGUUCCCAAAAGGGCGAACCAUCCGGAGUCCGGACGGCCAGCAUGUCUAGACUCGCUCCAGCCCCUAUACCAAAAAUAACACCGAAAAUACAG --.(((((.(((---------.((((((((((......)))))))))).)))(((((..(((((....).))))..)))))((....))......))))).................... ( -35.30) >DroSec_CAF1 17390 109 + 1 --CUUGGCUUGG---------UUGGUGCGUUCCCAAAAGGGCGAACCAUCCGGAGUCCGGACGGCCAGCAUGUCUAGACUCGCUCCAGCCCCUAUACCAAAAAUAACACCGAAAAUACAG --.((((...((---------((((.((((((((.....)).))))......(((((..(((((....).))))..))))))).))))))......)))).................... ( -34.30) >DroSim_CAF1 17578 109 + 1 --CGUGGCUUGG---------UUGGUUCGUUCCCAAAAGGGCGAACCAUCCGGAGUCCGGACGGCCAGCAUGUCUAGACUCGCUCCAGCCCCUAUACCAAAAAUAACACCGAAAAUACAG --.(.(((((((---------.((((((((((......)))))))))).)))(((((..(((((....).))))..))))).....)))).)............................ ( -36.10) >DroEre_CAF1 19443 109 + 1 --CUUGGUUUGG---------UUGGUUCGUUCCCAAAAGGGCGAACCAUCCGGAGUCCGGACGGCCAGCAUGUCUAGACUCGCUCCAGCCCCUAUACCAAAAAUAACACCGAAAAUACAG --.(((((.(((---------.((((((((((......)))))))))).)))(((((..(((((....).))))..)))))((....))......))))).................... ( -35.30) >DroYak_CAF1 19812 120 + 1 CUCUUGGUUUGGUUGGUUUGGUUGAUUCGGUCCCAAAAGGGCGAACCAUCCGGAGUCCGGACGGCCAGCAUGUCUAGACUCGCUCCAGCCCCUAUACCAAAAAUAACACCGAAAAUACAG ...(((((..((((((..(((.((.((((..(((....))))))).)).)))(((((..(((((....).))))..)))))...)))))).....))))).................... ( -37.10) >consensus __CUUGGUUUGG_________UUGGUUCGUUCCCAAAAGGGCGAACCAUCCGGAGUCCGGACGGCCAGCAUGUCUAGACUCGCUCCAGCCCCUAUACCAAAAAUAACACCGAAAAUACAG .....((((.((..........(((((((..(((....))))))))))....(((((..(((((....).))))..)))))...)))))).............................. (-30.16 = -30.32 + 0.16)



| Location | 12,080,667 – 12,080,776 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.18 |
| Mean single sequence MFE | -40.72 |
| Consensus MFE | -36.20 |
| Energy contribution | -36.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981722 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12080667 109 - 27905053 CUGUAUUUUCGGUGUUAUUUUUGGUAUAGGGGCUGGAGCGAGUCUAGACAUGCUGGCCGUCCGGACUCCGGAUGGUUCGCCCUUUUGGGAACGAACCAA---------CCAAACCAAG-- (((......)))........(((((...(((((((((.((.(.((((.....)))))))))))).))))((.((((((((((....)))..))))))).---------))..))))).-- ( -41.20) >DroSec_CAF1 17390 109 - 1 CUGUAUUUUCGGUGUUAUUUUUGGUAUAGGGGCUGGAGCGAGUCUAGACAUGCUGGCCGUCCGGACUCCGGAUGGUUCGCCCUUUUGGGAACGCACCAA---------CCAAGCCAAG-- (((......)))........(((((....((..(((.(((..((((((...((..(((((((((...)))))))))..))...))))))..))).))).---------))..))))).-- ( -39.90) >DroSim_CAF1 17578 109 - 1 CUGUAUUUUCGGUGUUAUUUUUGGUAUAGGGGCUGGAGCGAGUCUAGACAUGCUGGCCGUCCGGACUCCGGAUGGUUCGCCCUUUUGGGAACGAACCAA---------CCAAGCCACG-- (((......))).........((((...(((((((((.((.(.((((.....)))))))))))).))))((.((((((((((....)))..))))))).---------))..))))..-- ( -40.30) >DroEre_CAF1 19443 109 - 1 CUGUAUUUUCGGUGUUAUUUUUGGUAUAGGGGCUGGAGCGAGUCUAGACAUGCUGGCCGUCCGGACUCCGGAUGGUUCGCCCUUUUGGGAACGAACCAA---------CCAAACCAAG-- (((......)))........(((((...(((((((((.((.(.((((.....)))))))))))).))))((.((((((((((....)))..))))))).---------))..))))).-- ( -41.20) >DroYak_CAF1 19812 120 - 1 CUGUAUUUUCGGUGUUAUUUUUGGUAUAGGGGCUGGAGCGAGUCUAGACAUGCUGGCCGUCCGGACUCCGGAUGGUUCGCCCUUUUGGGACCGAAUCAACCAAACCAACCAAACCAAGAG ..........(((.......(((((...(((((((((.((.(.((((.....)))))))))))).))))((.((((((((((....)))..))))))).))..)))))....)))..... ( -41.00) >consensus CUGUAUUUUCGGUGUUAUUUUUGGUAUAGGGGCUGGAGCGAGUCUAGACAUGCUGGCCGUCCGGACUCCGGAUGGUUCGCCCUUUUGGGAACGAACCAA_________CCAAACCAAG__ (((......))).......((((((...(((((((((.((.(.((((.....)))))))))))).))))...((((((((((....)))..)))))))..............)))))).. (-36.20 = -36.20 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:47 2006