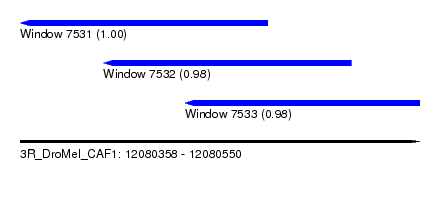
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,080,358 – 12,080,550 |
| Length | 192 |
| Max. P | 0.999850 |

| Location | 12,080,358 – 12,080,477 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.15 |
| Mean single sequence MFE | -31.07 |
| Consensus MFE | -30.56 |
| Energy contribution | -30.56 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.98 |
| SVM decision value | 4.25 |
| SVM RNA-class probability | 0.999850 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
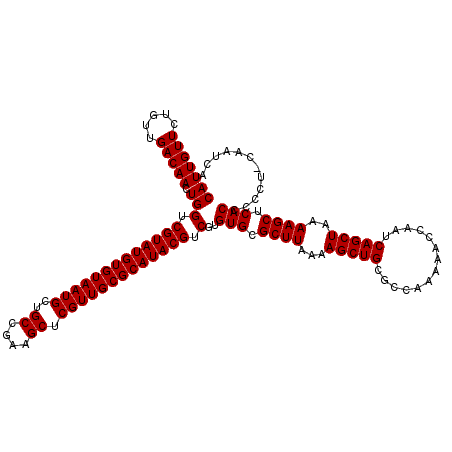
>3R_DroMel_CAF1 12080358 119 - 27905053 GAAGCUCGUUGCGCAUACGUCGUGUGCGCUUAAAAGCUGCGCUAAAAACCAAUCAGCUAAAAGCUCACCCCCU-CAAUCAAAUCAAACCCCAUCAGCUGAGCUUCGCAACGGGAAAACCA (((((((((((.((((((...))))))((((...(((((..............)))))..)))).........-...................)))).))))))).....((.....)). ( -30.74) >DroSec_CAF1 17081 119 - 1 GAAGCUCGUUGCGCAUACGUCGUGUGCGCUUAAAAGCUGCGCCAAAAACCAAUCAGCUAAAAGCUCACCCCCU-CAAUCGAAUCAAACCCCAUCAGCUGAGCUUCGCAACGGGAAAACCA (((((((...((((((((...)))))))).....(((((...............(((.....))).......(-(....))............)))))))))))).....((.....)). ( -31.90) >DroSim_CAF1 17269 119 - 1 GAAGCUCGUUGCGCAUACGUCGUGUGCGCUUAAAAGCUGCGCCAAAAACCAAUCAGCUAAAAGCUCACCCCCU-CAAUCAAAUCAAACCCCAUCAGCUGAGCUUCGCAACGGGAAAACCA (((((((((((.((((((...))))))((((...(((((..............)))))..)))).........-...................)))).))))))).....((.....)). ( -30.74) >DroEre_CAF1 19133 119 - 1 GAAGCUCGUUGCGCAUACGUCGUGUGCGCUUAAAAGCUGCGCCAAAAACCAAUCAGCUAAAAGCUCACCCCCC-CAAUCAAAUCAAAUCGCAUCAGCUGAGCUUCGCAACGGGAAAACCA ((((((((((((((((((...))))))((((...(((((..............)))))..)))).........-...............)))...)).))))))).....((.....)). ( -30.84) >DroYak_CAF1 19494 120 - 1 GAAGCUCGUUGCGCAUACGUCGUGUGCGCUUAAAAGCUGCGCCAAAAACCAAUCAGCUAAAAGCUCACCCCCCUCAAUCAAAUCAAAUCGCAUCAGCUGAGCUUCGCAACGGGAAAACCC ((((((((((((((((((...))))))((((...(((((..............)))))..)))).........................)))...)).))))))).....(((....))) ( -31.14) >consensus GAAGCUCGUUGCGCAUACGUCGUGUGCGCUUAAAAGCUGCGCCAAAAACCAAUCAGCUAAAAGCUCACCCCCU_CAAUCAAAUCAAACCCCAUCAGCUGAGCUUCGCAACGGGAAAACCA (((((((((((.((((((...))))))((((...(((((..............)))))..)))).............................)))).))))))).....((.....)). (-30.56 = -30.56 + 0.00)



| Location | 12,080,398 – 12,080,517 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.65 |
| Mean single sequence MFE | -34.83 |
| Consensus MFE | -31.64 |
| Energy contribution | -31.64 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.976080 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12080398 119 - 27905053 CAUUGUUCUGUUGACAACUGGUCGUAUGUGUAAUGCUGCCGAAGCUCGUUGCGCAUACGUCGUGUGCGCUUAAAAGCUGCGCUAAAAACCAAUCAGCUAAAAGCUCACCCCCU-CAAUCA .........(((((......(.(((((((((((((..((....)).))))))))))))).)..(((.((((...(((((..............)))))..)))).)))....)-)))).. ( -33.14) >DroSec_CAF1 17121 119 - 1 CAUUGUUCUGUUGACAACUGGUCGUAUGUGUAAUGCUGCCGAAGCUCGUUGCGCAUACGUCGUGUGCGCUUAAAAGCUGCGCCAAAAACCAAUCAGCUAAAAGCUCACCCCCU-CAAUCG .........(((((......(.(((((((((((((..((....)).))))))))))))).)..(((.((((...(((((..............)))))..)))).)))....)-)))).. ( -33.14) >DroSim_CAF1 17309 119 - 1 CAUUGUUCUGUUGACAACUGGUCGUAUGUGUAAUGCUGCCGAAGCUCGUUGCGCAUACGUCGUGUGCGCUUAAAAGCUGCGCCAAAAACCAAUCAGCUAAAAGCUCACCCCCU-CAAUCA .........(((((......(.(((((((((((((..((....)).))))))))))))).)..(((.((((...(((((..............)))))..)))).)))....)-)))).. ( -33.14) >DroEre_CAF1 19173 119 - 1 CAUUGUUCUGGUGACAACUGGUCGUAUGUGUAAUGCAGCCGAAGCUCGUUGCGCAUACGUCGUGUGCGCUUAAAAGCUGCGCCAAAAACCAAUCAGCUAAAAGCUCACCCCCC-CAAUCA .((((....(((((....(((((((((((((((((.(((....))))))))))))))))..(.(((((((....))).)))))....))))....((.....)))))))....-)))).. ( -39.40) >DroYak_CAF1 19534 120 - 1 CAUUGUUCUGUUGACAACUGGUCGUAUGUGUAAUGCAGCCGAAGCUCGUUGCGCAUACGUCGUGUGCGCUUAAAAGCUGCGCCAAAAACCAAUCAGCUAAAAGCUCACCCCCCUCAAUCA .........(((((......(.(((((((((((((.(((....)))))))))))))))).)..(((.((((...(((((..............)))))..)))).))).....))))).. ( -35.34) >consensus CAUUGUUCUGUUGACAACUGGUCGUAUGUGUAAUGCUGCCGAAGCUCGUUGCGCAUACGUCGUGUGCGCUUAAAAGCUGCGCCAAAAACCAAUCAGCUAAAAGCUCACCCCCU_CAAUCA (((((((.....))))).))(.(((((((((((((..((....)).))))))))))))).)..(((.((((...(((((..............)))))..)))).)))............ (-31.64 = -31.64 + 0.00)


| Location | 12,080,437 – 12,080,550 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.85 |
| Mean single sequence MFE | -40.58 |
| Consensus MFE | -36.16 |
| Energy contribution | -35.92 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.982511 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
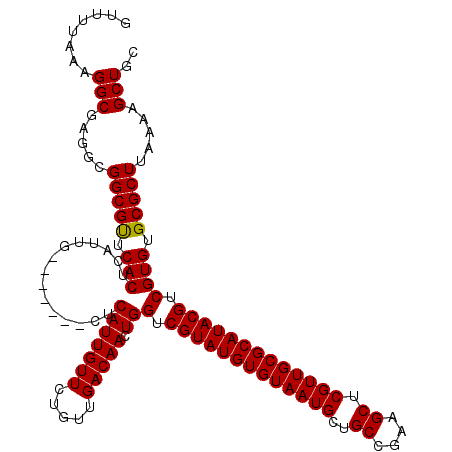
>3R_DroMel_CAF1 12080437 113 - 27905053 GUUUUAAAGGCGAGGCGGCGUUCACUCAUUG-------CUCAUUGUUCUGUUGACAACUGGUCGUAUGUGUAAUGCUGCCGAAGCUCGUUGCGCAUACGUCGUGUGCGCUUAAAAGCUGC (((((...(((..((((((((((((..((.(-------((..(((((.....)))))..))).))..))).)))))))))...)))....((((((((...))))))))...)))))... ( -40.70) >DroSec_CAF1 17160 113 - 1 GUUUUAAAGGCGAGGCGGCGUCCACUCAUUG-------CUCAUUGUUCUGUUGACAACUGGUCGUAUGUGUAAUGCUGCCGAAGCUCGUUGCGCAUACGUCGUGUGCGCUUAAAAGCUGC (((((...(((..((((((((.(((..((.(-------((..(((((.....)))))..))).))..)))..))))))))...)))....((((((((...))))))))...)))))... ( -37.90) >DroSim_CAF1 17348 113 - 1 GUUUUAAAGGCGAGGCGGCGUUCACUCAUUG-------CUCAUUGUUCUGUUGACAACUGGUCGUAUGUGUAAUGCUGCCGAAGCUCGUUGCGCAUACGUCGUGUGCGCUUAAAAGCUGC (((((...(((..((((((((((((..((.(-------((..(((((.....)))))..))).))..))).)))))))))...)))....((((((((...))))))))...)))))... ( -40.70) >DroEre_CAF1 19212 113 - 1 GUUUUAAAGGCGAGGCGGCGCUCACUCAUUG-------CUCAUUGUUCUGGUGACAACUGGUCGUAUGUGUAAUGCAGCCGAAGCUCGUUGCGCAUACGUCGUGUGCGCUUAAAAGCUGC ........(((.....(((((.(((.(((((-------.(((((.....)))))))).))(.(((((((((((((.(((....)))))))))))))))).)))).))))).....))).. ( -42.50) >DroYak_CAF1 19574 120 - 1 GUUUUAAAGGCGAGGCGGCGCUCACUCAUUGCUCAUUGCUCAUUGUUCUGUUGACAACUGGUCGUAUGUGUAAUGCAGCCGAAGCUCGUUGCGCAUACGUCGUGUGCGCUUAAAAGCUGC ........(((.....(((((.(((.....((.....)).(((((((.....))))).))(.(((((((((((((.(((....)))))))))))))))).)))).))))).....))).. ( -41.10) >consensus GUUUUAAAGGCGAGGCGGCGUUCACUCAUUG_______CUCAUUGUUCUGUUGACAACUGGUCGUAUGUGUAAUGCUGCCGAAGCUCGUUGCGCAUACGUCGUGUGCGCUUAAAAGCUGC ........(((.....(((((.(((...............(((((((.....))))).))(.(((((((((((((..((....)).))))))))))))).)))).))))).....))).. (-36.16 = -35.92 + -0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:45 2006